The Daffodil DNA Project: Results

Sample Naming
| 22FAlama: | Year |
| 22FAlama: | 2 character code representing the school: Forfar Academy |
| 22FAlama: | 4 character code indicating the variety: Lady Margaret |
Schools
| BA | Banchory Academy |
| BC | Beaulieu Convent School |
| CH | Cheam High School |
| CO | Combined |
| CM | Cockermouth School |
| EM | Emmanuel School |
| HT | Hautlieu |
| LA | Lockerbie Academy |
| LC | London Calling |
| LS | Luton Sixth Form College |
| FA | Forfar Academy |
| MA | Morgan Academy |
| QA | Queen Anne High School |
| RM | Royal Masonic School for Girls |
| SA | St Mary's Music School |
| SM | St Modan's High School |
| SP | St Peter the Apostle High School |
| TA | St Thomas of Aquin's High School |
| MG | St. Margaret's School for Girls |
| MY | Mearns Academy |
| MS | Morrison Academy |
| WS | Websters School |
| WH | Westray High School |
Varieties
| cher | Cheerio |
| coul | Coulmony |
| cove | Coverack Beauty |
| empr | Empress |
| fofi | Forest Fire |
| furn | Furness |
| lama | Lady Margaret Boscawen |
| lofy | Loch Fyne |
| luci | Lucifer |
| mihu | Minnie Hume |
| morv | Morven |
| orna | Ornatus |
| poet | Poetarum |
| prin | Princeps |
| alba | Albatross |
| tazz | Tezzeta |
| seal | Sealing Wax |
| swee | Sweetness |
| duma | Dutch Master |
| hiso | High Society |
| reds | Redstart |
| topo | Topolino |
| carl | Carlton |
| lown | Lowen |
| fern | Ferness |
| ther | Therapia |
| cobo | Copper Bowl |
| rocu | Rose of Cuan |
| cast | Castermoon |
| tahi | Tahiti |
| coma | Cornish May |
| coni | Cornish Niveth |
| gapl | Galanthus plicatus |
| phey | Pheasants Eye |
| duca | Ducat |
| goec | Golden Echo |
| mart | Martinette |
| verd | Verdin |
| prom | Prom Dance |
| germ | Geranium |
| fort | Fortune |
| seav | Sempre Avanti |
| lofi | Loch Fyne |
| tenb | Tenby |
| tete | Tete-a-tete |
| minn | Minnow |
| word | Wordsworth |
| pipr | Pink Pride |
| salo | Salome |
| redv | Red Devon |
Data Usage Policy
The data available from this site have been generated as part of a Royal Society (London) project (The Scottish Daffodil Project) to introduce 16-18 year old school pupils across Scotland to genome sequencing, molecular evolution and ecology.
The data are all freely available for teaching and research but are preliminary prior to formal publication by the Scottish Daffodil Project. The Scottish Daffodil Project is preparing publications that include: (a) the principals and teaching value of practical genome sequencing and genome analysis in schools and (b) the scientific interpretation of the genome data generated in the Project.
The expectation is that the Scottish Daffodil Project will submit articles within 18 months of the first release of raw data. If you wish to use the data in your own publications during this time, then please contact the Scottish Daffodil Project to ensure you have the best quality data to work from. Also, the following acknowledgment should be included: "These data were produced by the Scottish Daffodil Project in collaboration." We request that you notify the Scottish Daffodil Project upon publication so that this information can be included in the final annotation of the data and reporting on the Scottish Daffodil Project.
While still in waiting period status, the assembly and raw sequence reads should not be redistributed or repackaged without permission from Scottish Daffodil Project." Once moved to unreserved status, the data are freely available for any subsequent use.
This project is a collaboration between Jon Hale (Head of Biology at Beaulieu Convent School, Jersey), the University of Dundee School of Life Sciences and the James Hutton Institute.
Schools are working in conjunction with STEM partners from the University of Dundee and the James Hutton Institute to sample various daffodil varieties and carry out DNA sequencing of the chloroplast genome using Oxford Nanopore MinIon sequencing. The schools are then carrying out analysis of the resulting sequence to answer specific research questions.
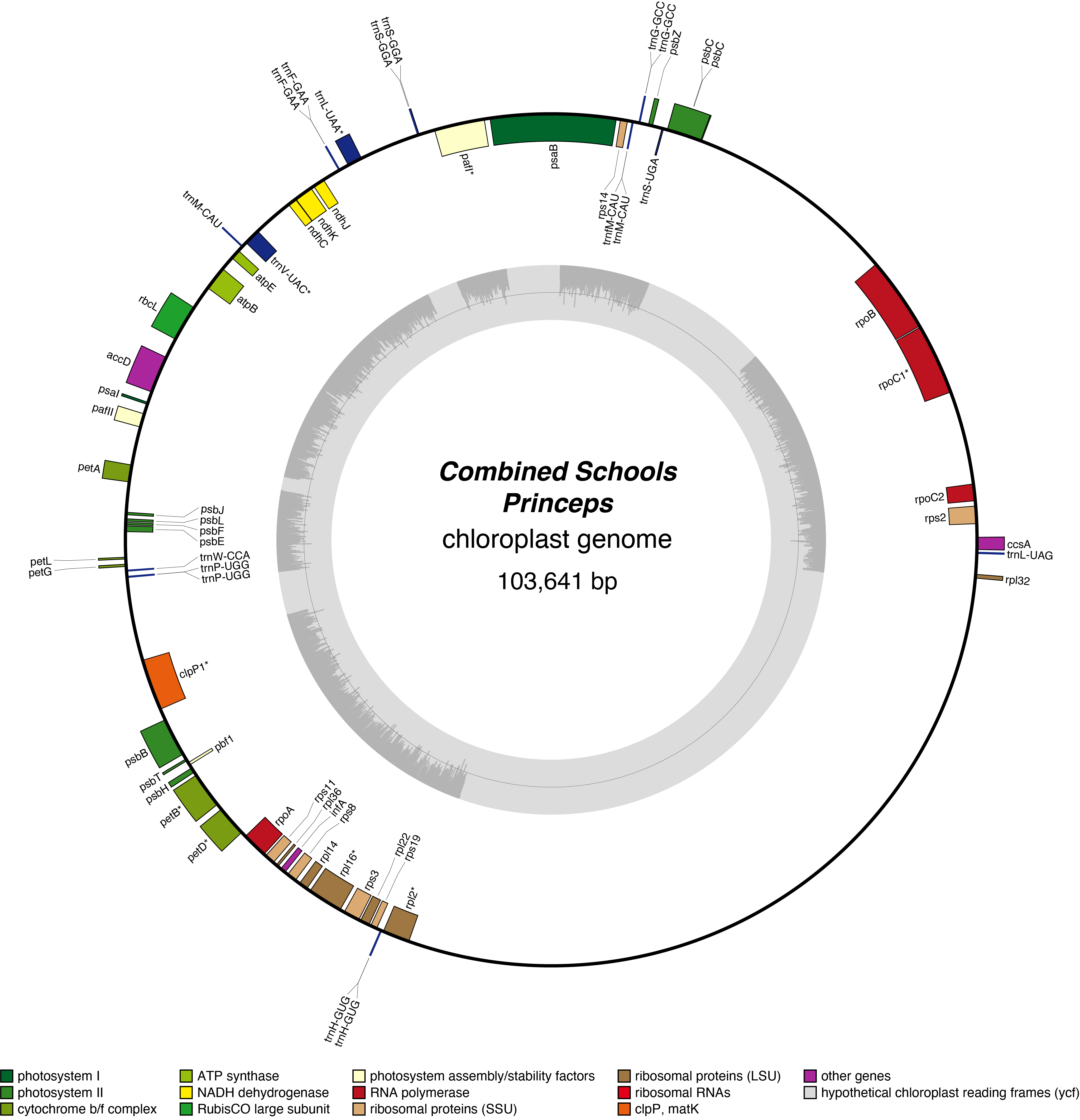
The resulting sequencing data from all the schools are being collated on this site, allowing with additional sequences obtained by Beaulieu Convent School, Jersey. This enables the phylogeny of the various varietites to be inferred.
This project has been funded by The Royal Society, with additional funding from the Friends of Dundee University Botanic Garden to enable the University of Dundee Data Analysis Group to provide this centralised resource with consistent analysis processes.
We would be happy for anyone to reuse this data in their own projects, but please see our Data Usage Policy.
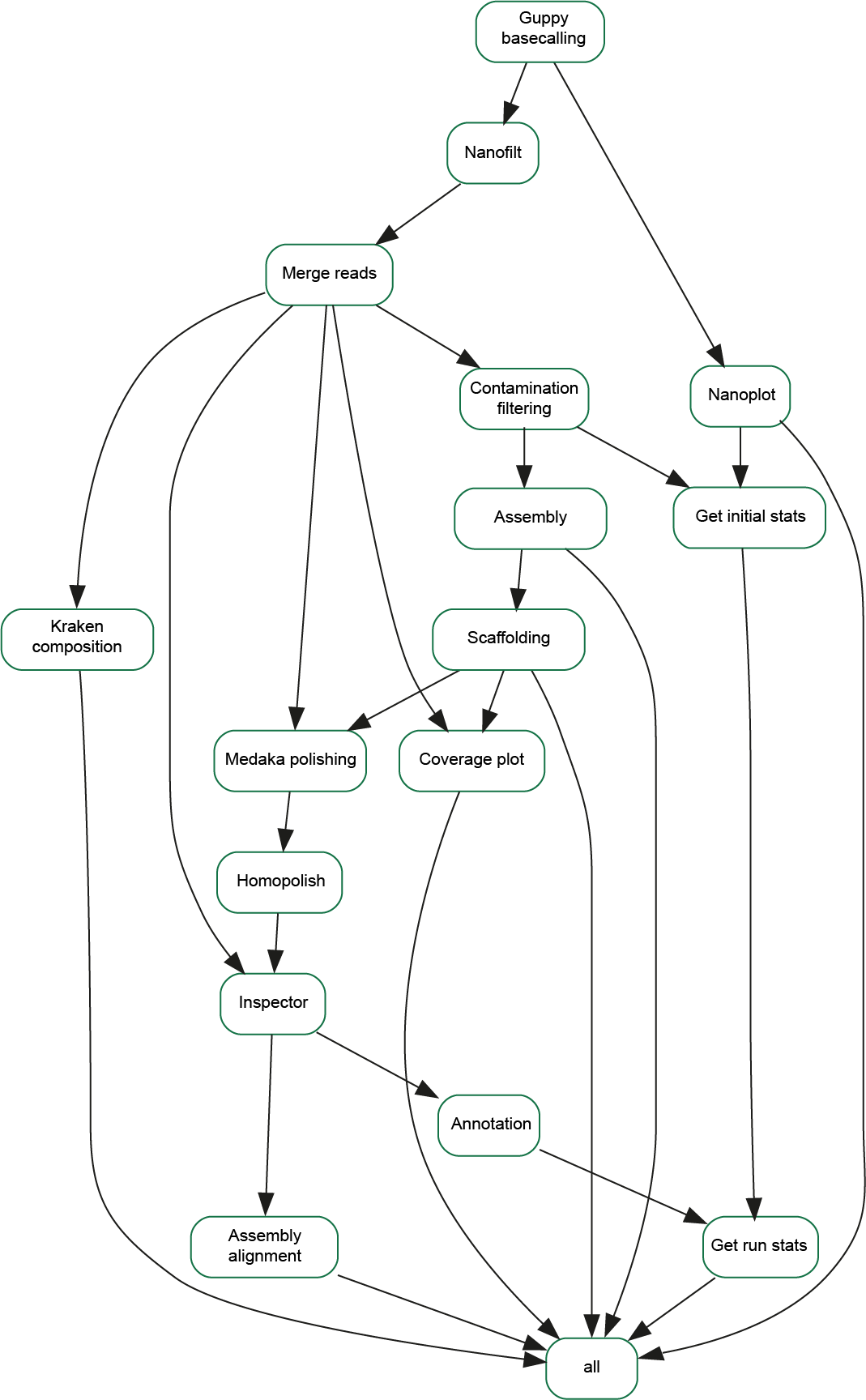
The sequences have all been processed using an automated workflow consisting of a number of stages allowing us to get from the raw data produced by the MinIon sequencer to assembled, annotated sequences
Basecalling
This is the process of converting the signal from the MinIon sequencer into the DNA bases this represents. The instrument produces thousands of reads of varying length, up to 40-50kb in length. Basecalling was carried out using the Oxford Nanopore Guppy software (version 6.1.2) using the appropriate high accuracy model for the flowcells and sequencing kits used (dna_r9.4.1_450bps_hac).
Quality Filtering
Every base of sequence generated is assigned a quality score, which is a measure of how likely the base is to have been called correctly. These range from 0-40, using a logarithmic scale, where a score of 10 represents a 1 in 10 chance that the base call is incorrect, a score of 20 relating to a 1 in 100 chance, and 30 being 1 in 1000. Sequences are filtered using NanoFilt to remove sequences shorter than 300 bp and also those with a minimum average quality score below 10.
Contamination Filtering
A large proportion of these samples does not consist of daffodil chloroplastic DNA, so to make the assembly process easier, contaminants are removed from the sequence data. Kraken 2.1.2 is also used to identify what the origin of the seqeunces in each sample. The sequence reads are mapped to a known daffodil chloroplast sequence using winnowmap 2.03, and those which do not have similarity with this reference sequence are discarded.
Assembly
- nextDenovo is a type of assembler known as a string-graph assembler. This requires longer reads, and is capable of producing a fully intact chloroplast assembly given sufficient data with long enough reads. nextDenovo is able to correctly assemble the inverted repeats, so is used first with each data set. If insufficient data is available, the assembly will fail.
- Flye is used if nextDenovo is unable to assemble the data. Flye is a repeat-graph assembler, which works in a different way to nextDenovo. There is typically far more data present in these samples than required for assembly, so the longest reads are used, with enough reads to cover the genome 50 times. Once assembled, the remaining reads are then mapped against the assembly to try to identify and remove errors in a process called polishing. 3 iterations of polishing were carried out in this case.
Scaffolding
Orientation
Polishing
- Medaka is used first, which is a general polishing tool.
- Homopolish tries to correct a particular class of error which occurs when there is a run of the same base a number of times (known as homopolymer repeats). These can be a problem since it can be hard to determine quite how many times the base occurs, leading to expansion or compression of the repeat.
- The final tool (inspector) compares the assembly with the existing MH706763 assembly to try to identify any structural issues present.
Annotation
Multiple Sequence Alignment
Phylogeny
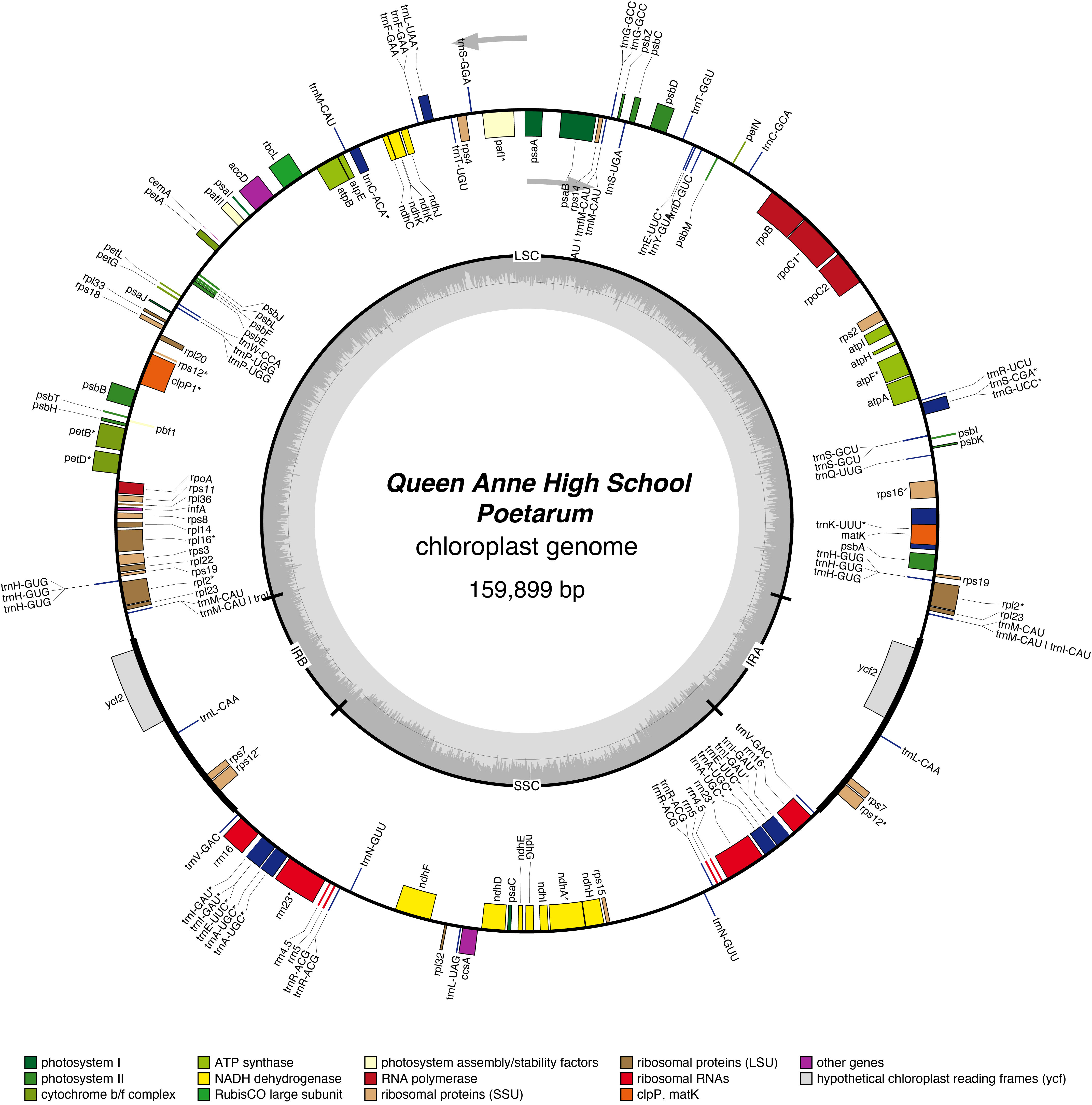
2025 Daffodils
Ducat Chloroplast Genome
- Corona: Yellow
- Perianth: Yellow
- Division 1 (Trumpet Daffodils)
- Registered: <1907
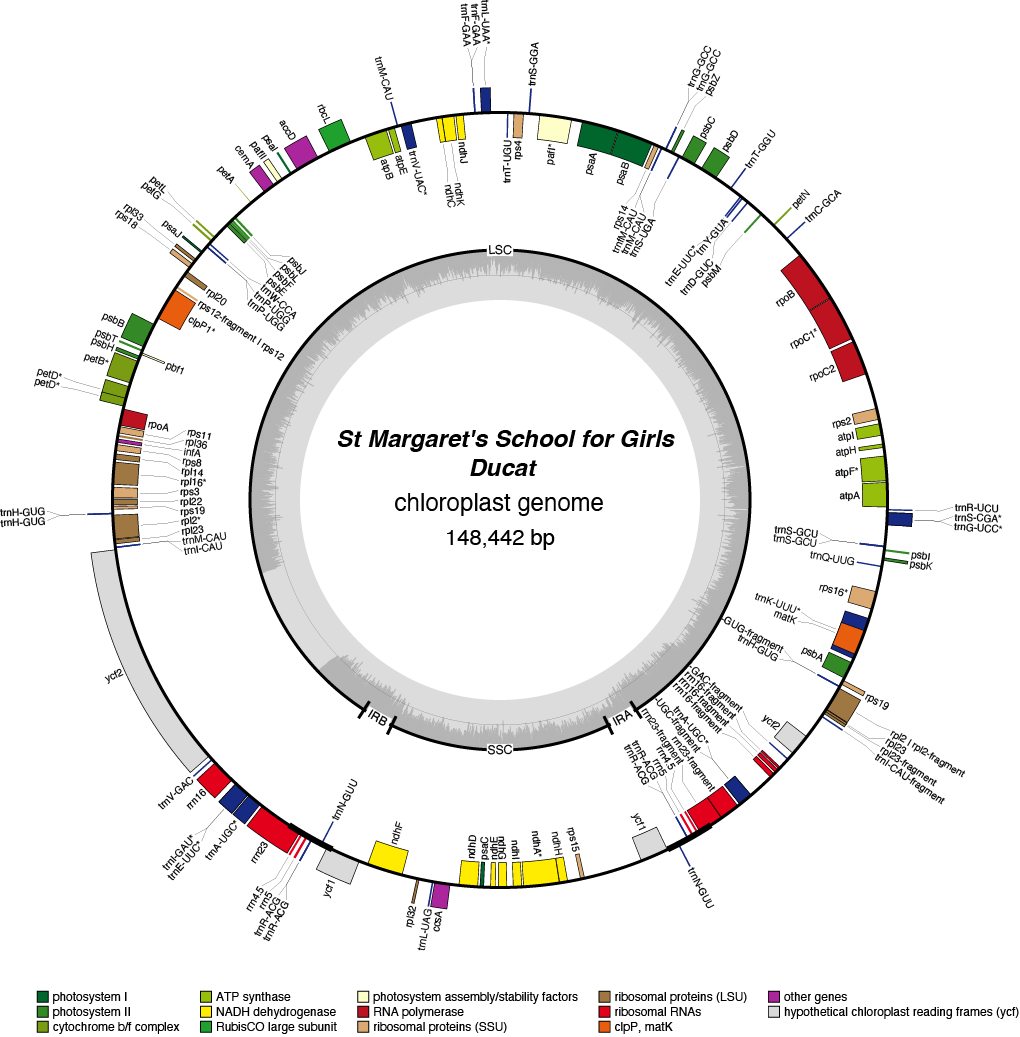
| Total Sequenced Bases (bp)
help
|
684,849,523 | Mean Quality
help
|
6.8 |
| Mean Read Length (bp)
help
|
1,010.5 | N50 Read Length (bp)
help
|
3,689.0 |
| Proportion of Chloroplast Bases (%)
help
|
3.5 | Chloroplast Coverage
help
|
148 |
| Assembled Contigs
help
|
5 | Assembled Contig Length (bp)
help
|
159524 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
148429 |
Dutch Master Chloroplast Genome
- Perianth: yellow
- Corona: yellow
- Division 7 (Jonquilla Daffodils)
- Hybridiser: Matthew Zandbergen
- Registered: 1948
| Total Sequenced Bases (bp)
help
|
322,659,260 | Mean Quality
help
|
11.9 |
| Mean Read Length (bp)
help
|
687.0 | N50 Read Length (bp)
help
|
1,102.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.48 | Chloroplast Coverage
help
|
9 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Fortune Chloroplast Genome
- Perianth: yellow
- Corona: orange
- Division 2 (Large cupped daffodils)
- Hybridiser: Walter T. Ware
- Registered: <1938
- Seed parent: Sir Watkin
- Pollen parent: Blackwell

| Total Sequenced Bases (bp)
help
|
130,361,761 | Mean Quality
help
|
13.3 |
| Mean Read Length (bp)
help
|
1,007.4 | N50 Read Length (bp)
help
|
1,733.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.63 | Chloroplast Coverage
help
|
5 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Fortune Chloroplast Genome
- Perianth: yellow
- Corona: orange
- Division 2 (Large cupped daffodils)
- Hybridiser: Walter T. Ware
- Registered: <1938
- Seed parent: Sir Watkin
- Pollen parent: Blackwell
| Total Sequenced Bases (bp)
help
|
22,232,987 | Mean Quality
help
|
10.7 |
| Mean Read Length (bp)
help
|
936.5 | N50 Read Length (bp)
help
|
1,666.0 |
| Proportion of Chloroplast Bases (%)
help
|
2.2 | Chloroplast Coverage
help
|
3 |
| Assembled Contigs
help
|
9 | Assembled Contig Length (bp)
help
|
52577 |
| Assembled Scaffolds
help
|
9 | Assembled Scaffold Length (bp)
help
|
52577 |
Geranium Chloroplast Genome

| Total Sequenced Bases (bp)
help
|
135,057,881 | Mean Quality
help
|
4.6 |
| Mean Read Length (bp)
help
|
1,043.2 | N50 Read Length (bp)
help
|
1,858.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.22 | Chloroplast Coverage
help
|
1 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
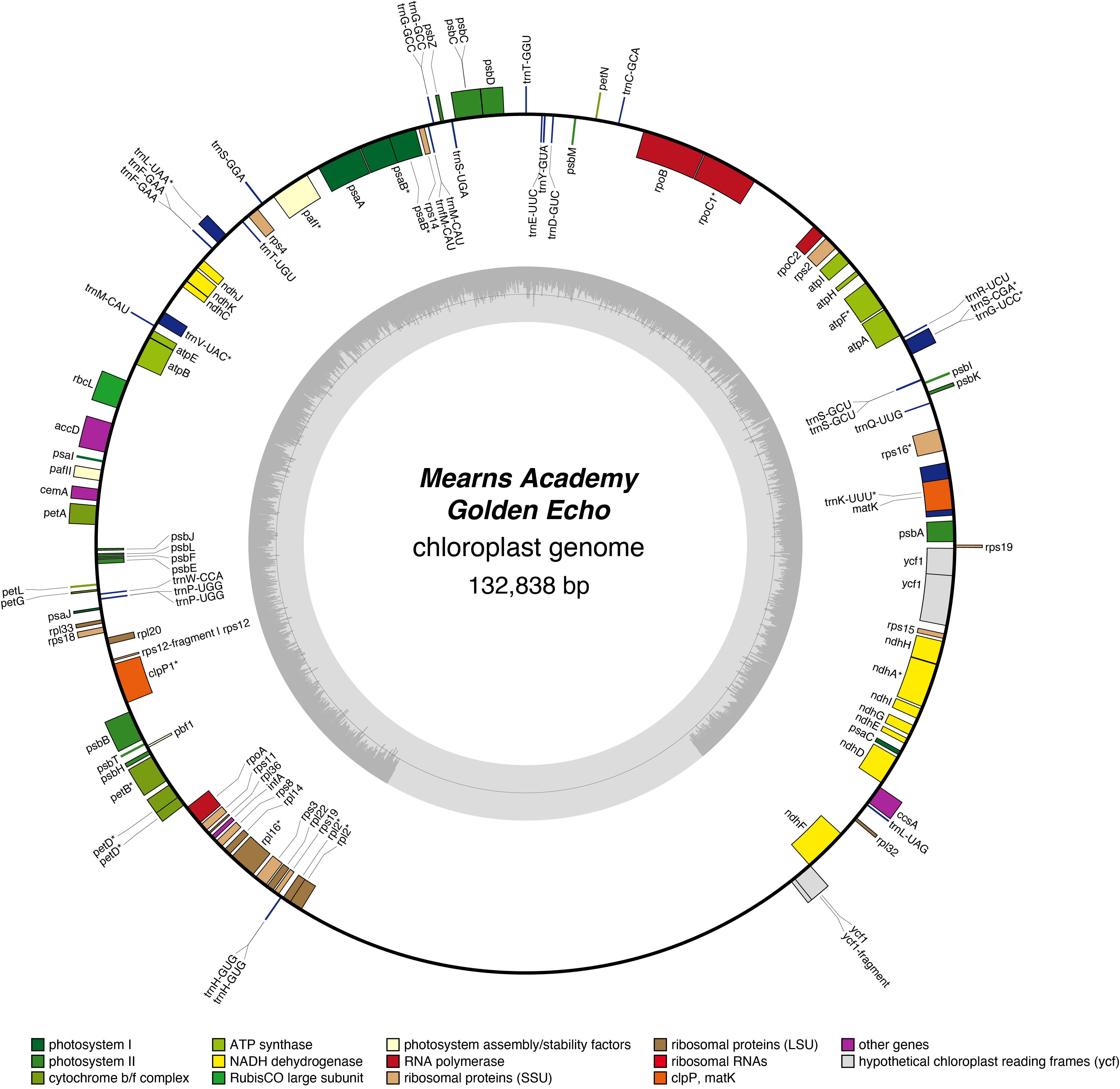
Golden Echo Chloroplast Genome
- Perianth: white, with yellow centre
- Corona: yellow
- Division 7 (Jonquilla Daffodils) eli>Hybridiser: Brent Heath
- Registered: 2014

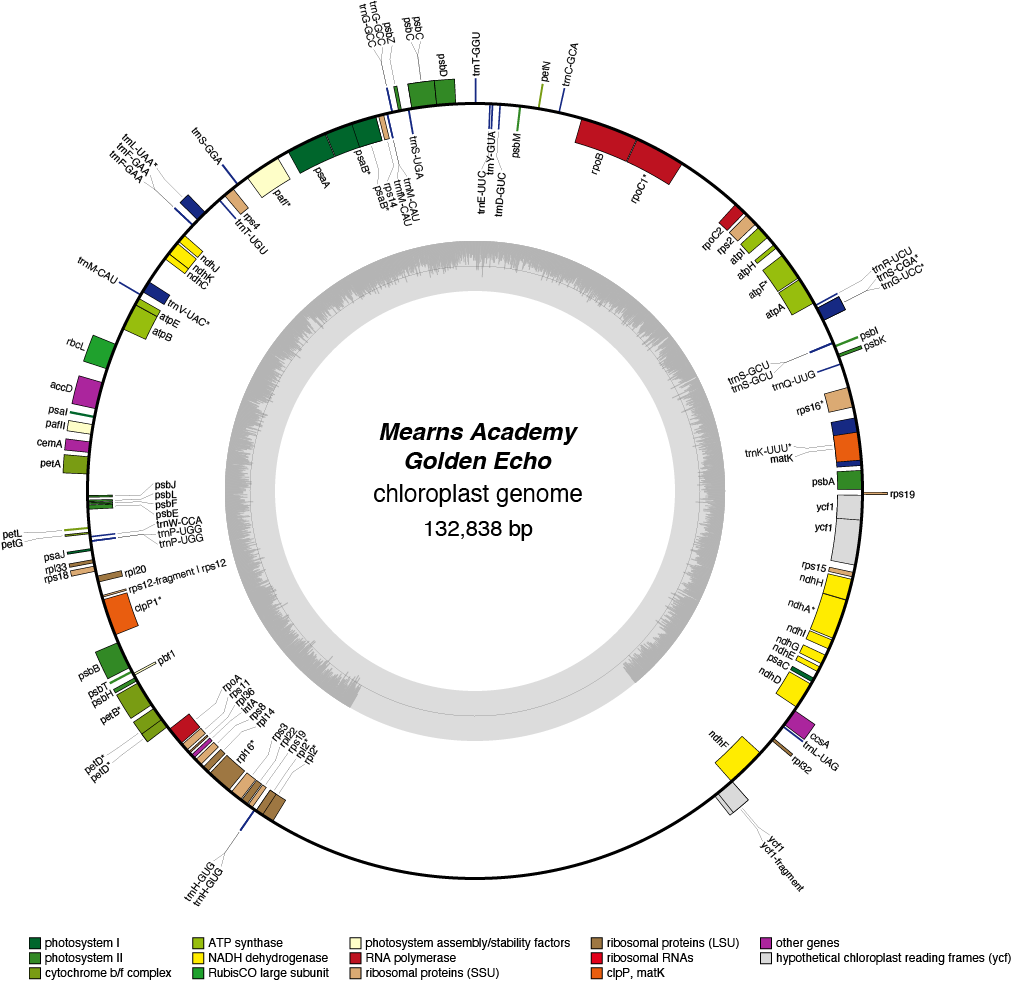
| Total Sequenced Bases (bp)
help
|
410,529,960 | Mean Quality
help
|
11.5 |
| Mean Read Length (bp)
help
|
1,362.3 | N50 Read Length (bp)
help
|
3,075.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.3 | Chloroplast Coverage
help
|
34 |
| Assembled Contigs
help
|
4 | Assembled Contig Length (bp)
help
|
106785 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
133035 |
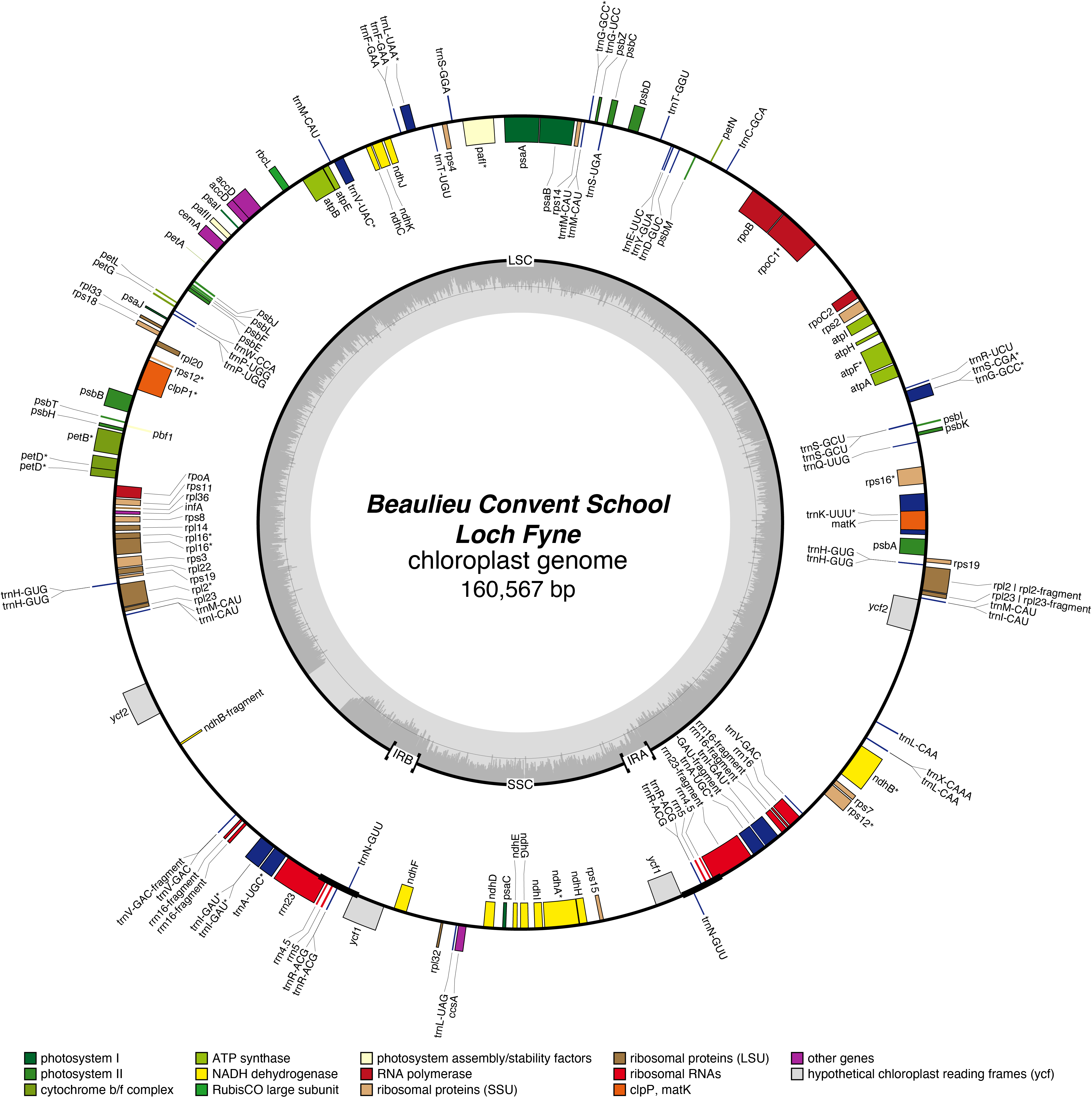
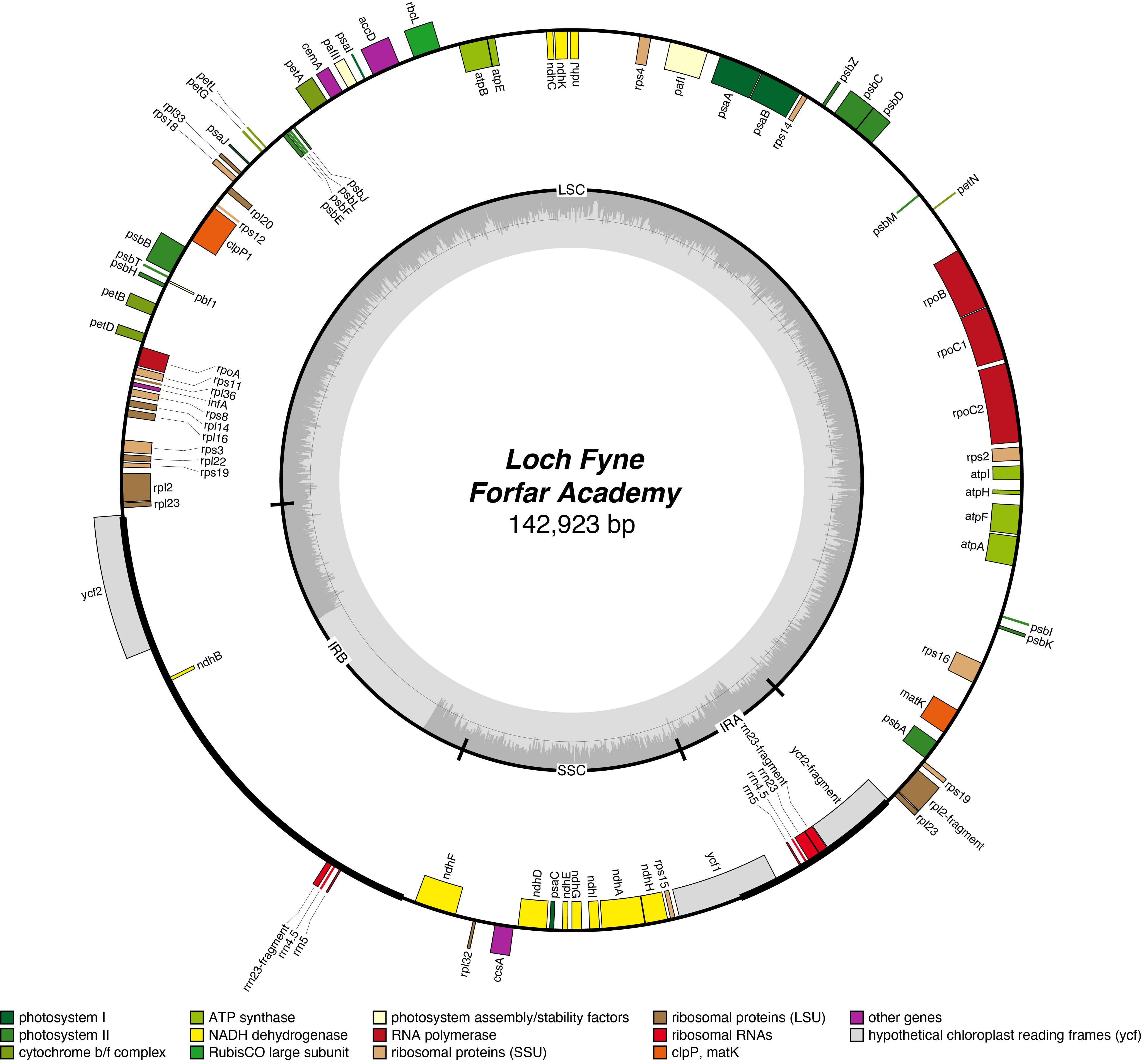
Loch Fyne Chloroplast Genome
- Corona: yellow
- Perianth: white
- Division: 2 (Large-Cupped Daffodils)
- Hybridiser: The Brodie of Brodie
- Registered: <1911

| Total Sequenced Bases (bp)
help
|
14,275,886 | Mean Quality
help
|
12.9 |
| Mean Read Length (bp)
help
|
1,270.9 | N50 Read Length (bp)
help
|
8,554.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.56 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Lowen Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
168,415,479 | Mean Quality
help
|
8.3 |
| Mean Read Length (bp)
help
|
1,853.4 | N50 Read Length (bp)
help
|
13,082.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.82 | Chloroplast Coverage
help
|
8 |
| Assembled Contigs
help
|
19 | Assembled Contig Length (bp)
help
|
75134 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
139228 |
Martinette Chloroplast Genome
- Perianth: yellow
- Corona: orange
- Division 8 (Tazetta Daffodils) eli>Hybridiser: Harry I. Tuggle Jr.
- Registered: 1985

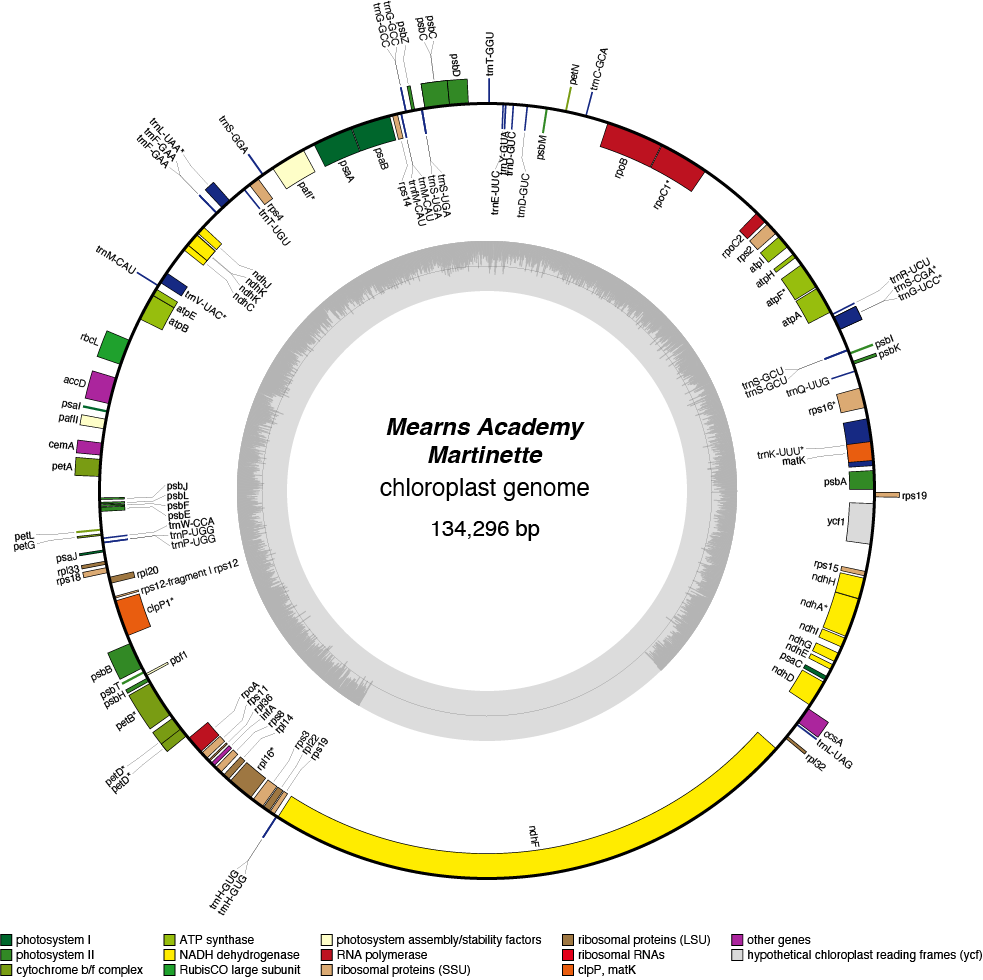
| Total Sequenced Bases (bp)
help
|
502,590,576 | Mean Quality
help
|
11.8 |
| Mean Read Length (bp)
help
|
905.7 | N50 Read Length (bp)
help
|
1,638.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.6 | Chloroplast Coverage
help
|
49 |
| Assembled Contigs
help
|
11 | Assembled Contig Length (bp)
help
|
107596 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
135157 |
Minnow Chloroplast Genome
- Perianth: white
- Corona: yellow
- Division 8 (Tazetta Daffodils)
- Hybridiser: Alec Gray
- Registered: 1962
| Total Sequenced Bases (bp)
help
|
37,678,013 | Mean Quality
help
|
7.9 |
| Mean Read Length (bp)
help
|
576.3 | N50 Read Length (bp)
help
|
15,914.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.055 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Pink Pride Chloroplast Genome
- Corona: Pink
- Perianth: white
- Division: 2 (large-cupped daffodils)
- Hybridiser: W.P. van Eeden
- Registered: 1970
| Total Sequenced Bases (bp)
help
|
289,927,776 | Mean Quality
help
|
13.6 |
| Mean Read Length (bp)
help
|
559.7 | N50 Read Length (bp)
help
|
826.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.55 | Chloroplast Coverage
help
|
10 |
| Assembled Contigs
help
|
9 | Assembled Contig Length (bp)
help
|
24836 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
229693 |
Prom Dance Chloroplast Genome
- Perianth: white
- Corona: yello
- Division 11a (Collar Daffodils) eli>Hybridiser: J.S.Pennings
- Registered: 2016
| Total Sequenced Bases (bp)
help
|
929,916,767 | Mean Quality
help
|
5.1 |
| Mean Read Length (bp)
help
|
1,590.6 | N50 Read Length (bp)
help
|
148,692.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.00011 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Red Devon Chloroplast Genome
- Perianth: yellow
- Corona: orange
- Division 2 (Large cupped daffodils)
- Hybridiser: E.B. Champernowne
- Registered: <1943
- Seed parent: Fortune
- Pollen parent: Killigrew
| Total Sequenced Bases (bp)
help
|
33,500,953 | Mean Quality
help
|
6.5 |
| Mean Read Length (bp)
help
|
989.3 | N50 Read Length (bp)
help
|
7,018.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.48 | Chloroplast Coverage
help
|
1 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Salome Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
114,173,922 | Mean Quality
help
|
10.9 |
| Mean Read Length (bp)
help
|
1,127.8 | N50 Read Length (bp)
help
|
1,674.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.89 | Chloroplast Coverage
help
|
6 |
| Assembled Contigs
help
|
11 | Assembled Contig Length (bp)
help
|
74447 |
| Assembled Scaffolds
help
|
11 | Assembled Scaffold Length (bp)
help
|
74447 |
Sempre Avanti Chloroplast Genome
- Perianth: yellow
- Corona: orange
- Division 2 (Large cupped daffodils)
- Hybridiser: De Graaff Brothers
- Registered: <1938
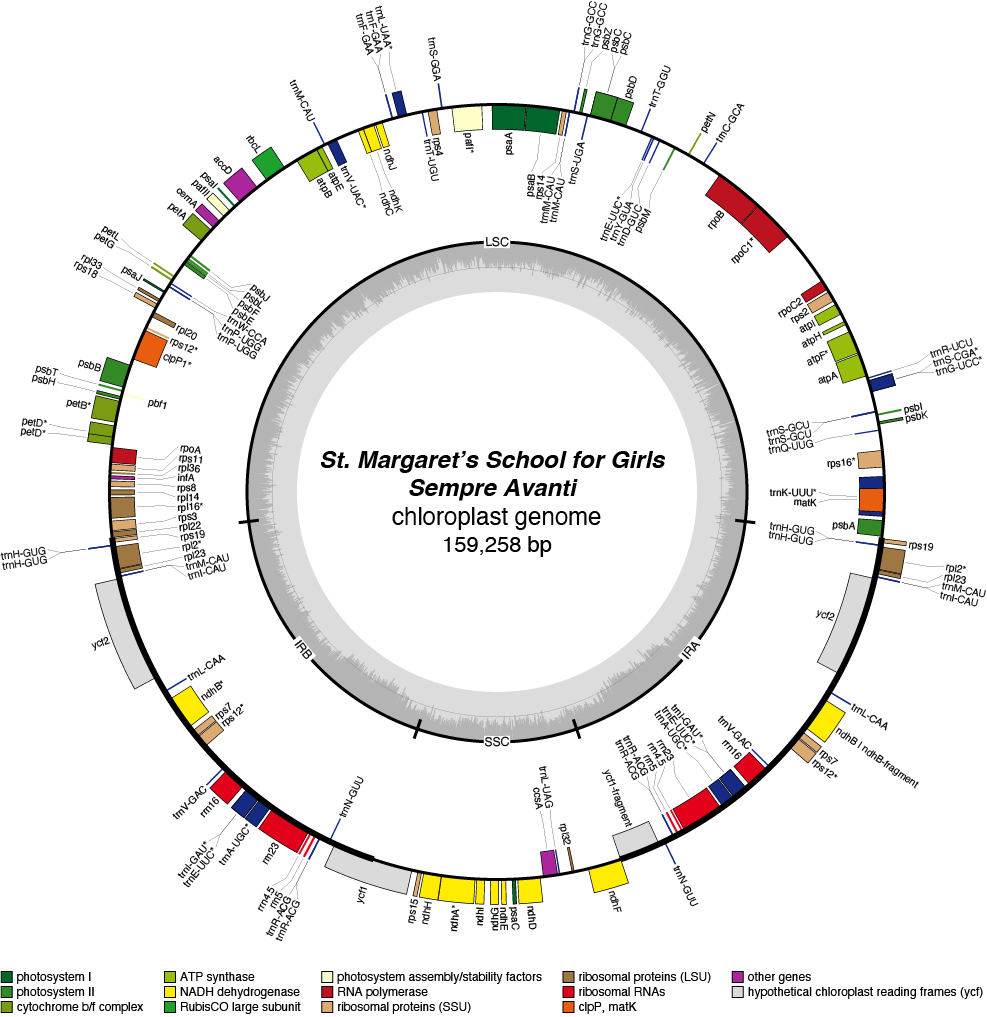
| Total Sequenced Bases (bp)
help
|
329,781,587 | Mean Quality
help
|
16.9 |
| Mean Read Length (bp)
help
|
2,339.8 | N50 Read Length (bp)
help
|
5,369.0 |
| Proportion of Chloroplast Bases (%)
help
|
4.7 | Chloroplast Coverage
help
|
96 |
| Assembled Contigs
help
|
6 | Assembled Contig Length (bp)
help
|
199269 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
347837 |
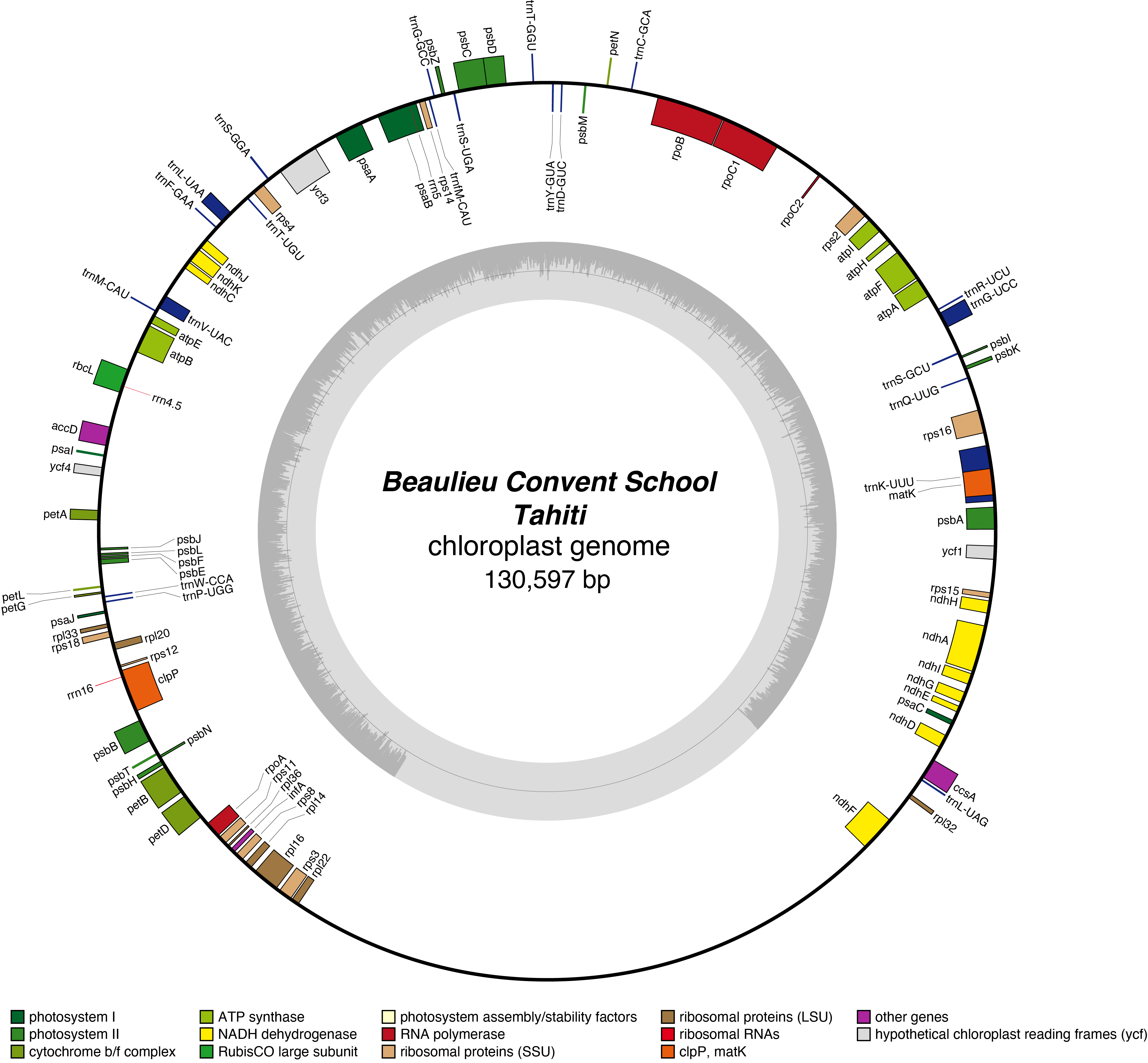
Tahiti Chloroplast Genome
- Corona: orange
- Perianth: yellow
- Division: 4 (Double daffodils)
- Hybridiser: J.L. Richardson
- Registered: 1956
| Total Sequenced Bases (bp)
help
|
100,747,589 | Mean Quality
help
|
14.1 |
| Mean Read Length (bp)
help
|
677.3 | N50 Read Length (bp)
help
|
1,280.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.087 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Tenby Chloroplast Genome
- Perianth: yellow
- Corona: yellow
- Division 13 (Wild Variants)

| Total Sequenced Bases (bp)
help
|
143,540,160 | Mean Quality
help
|
10.3 |
| Mean Read Length (bp)
help
|
901.3 | N50 Read Length (bp)
help
|
4,871.0 |
| Proportion of Chloroplast Bases (%)
help
|
1 | Chloroplast Coverage
help
|
9 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Tete-a-tete Chloroplast Genome
- Perianth: yellow
- Corona: yellow
- Division 12 (Miscellaneous)
- Hybridiser: Alec Gray
- Registered: <1949
- Seed parent: Cyclataz
- Pollen parent: unknown - openly pollenated
| Total Sequenced Bases (bp)
help
|
106,345,052 | Mean Quality
help
|
13.4 |
| Mean Read Length (bp)
help
|
996.2 | N50 Read Length (bp)
help
|
1,794.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.28 | Chloroplast Coverage
help
|
1 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Verdin Chloroplast Genome
- Perianth: yellow
- Corona: white
- Division 7 (Jonquilla Daffodils) eli>Hybridiser: Grant E. Mitsch
- Registered: 1965

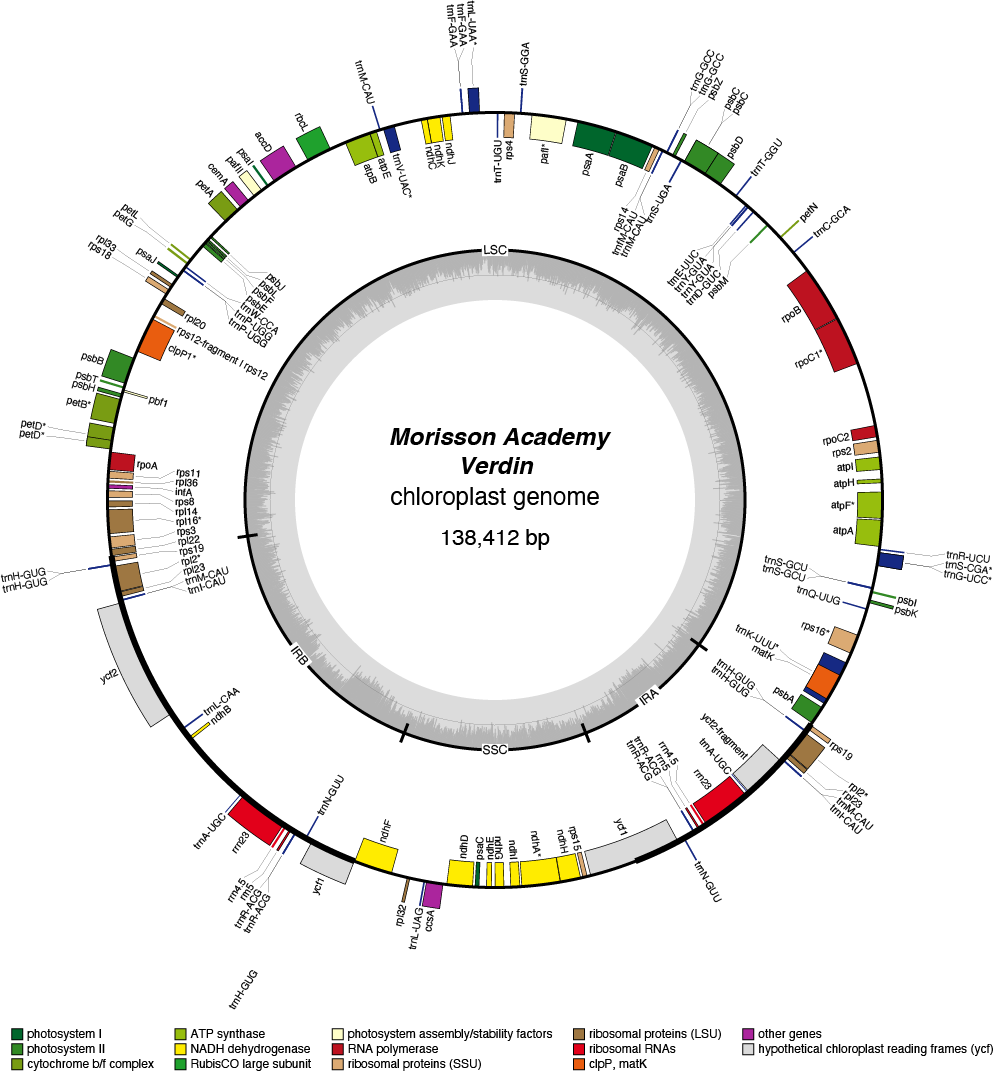
| Total Sequenced Bases (bp)
help
|
1,089,362,836 | Mean Quality
help
|
15.7 |
| Mean Read Length (bp)
help
|
2,471.4 | N50 Read Length (bp)
help
|
7,966.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.96 | Chloroplast Coverage
help
|
65 |
| Assembled Contigs
help
|
8 | Assembled Contig Length (bp)
help
|
175867 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
138428 |
Wordsworth Chloroplast Genome
- Perianth: white
- Corona: green-yellow-red
- Division 9 (Poeticus Daffodils)
- Hybridiser: William Balch
- Registered: <1931
| Total Sequenced Bases (bp)
help
|
841,138,166 | Mean Quality
help
|
17.9 |
| Mean Read Length (bp)
help
|
1,731.1 | N50 Read Length (bp)
help
|
2,991.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.43 | Chloroplast Coverage
help
|
22 |
| Assembled Contigs
help
|
10 | Assembled Contig Length (bp)
help
|
127187 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
133268 |
2024 Daffodils
Cornish Niveth Chloroplast Genome
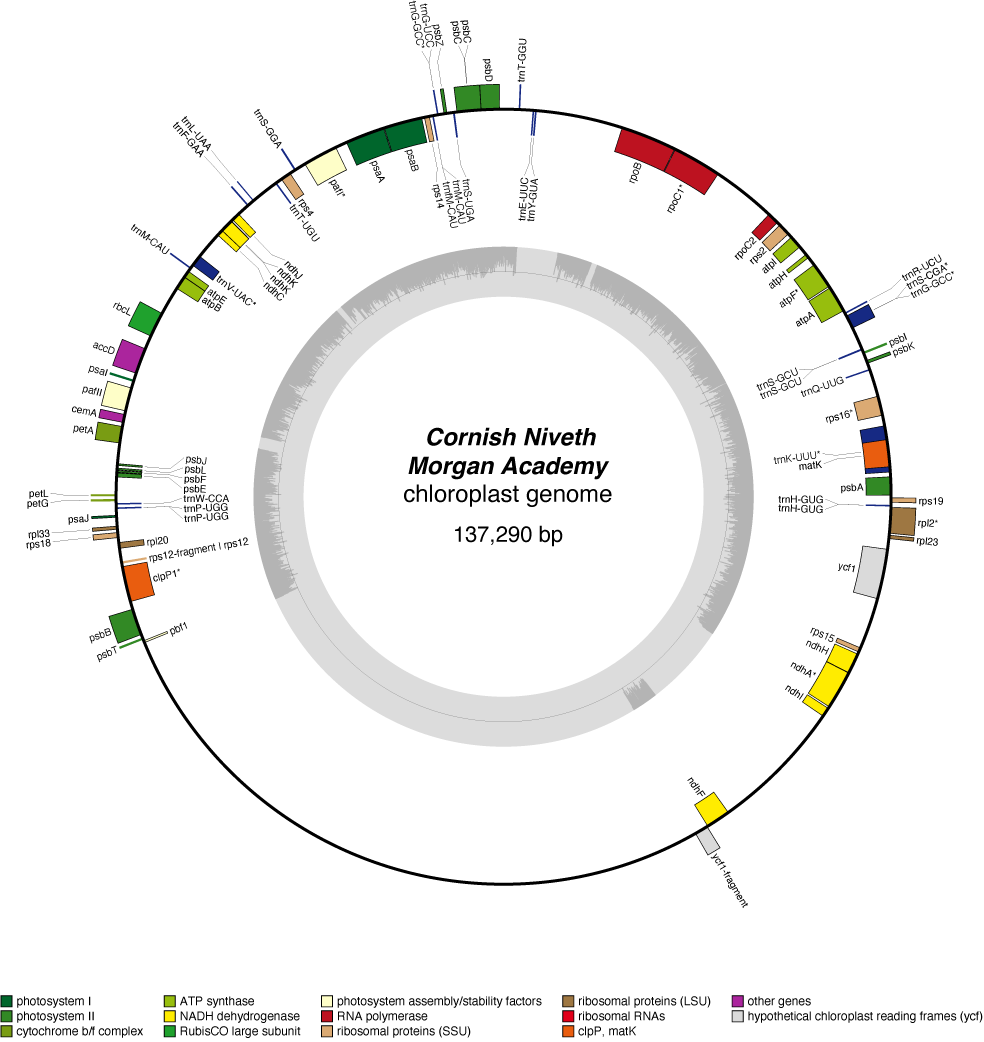
| Total Sequenced Bases (bp)
help
|
20,109,977 | Mean Quality
help
|
13.8 |
| Mean Read Length (bp)
help
|
1,723.5 | N50 Read Length (bp)
help
|
4,302.0 |
| Proportion of Chloroplast Bases (%)
help
|
3 | Chloroplast Coverage
help
|
3 |
| Assembled Contigs
help
|
8 | Assembled Contig Length (bp)
help
|
86996 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
137455 |
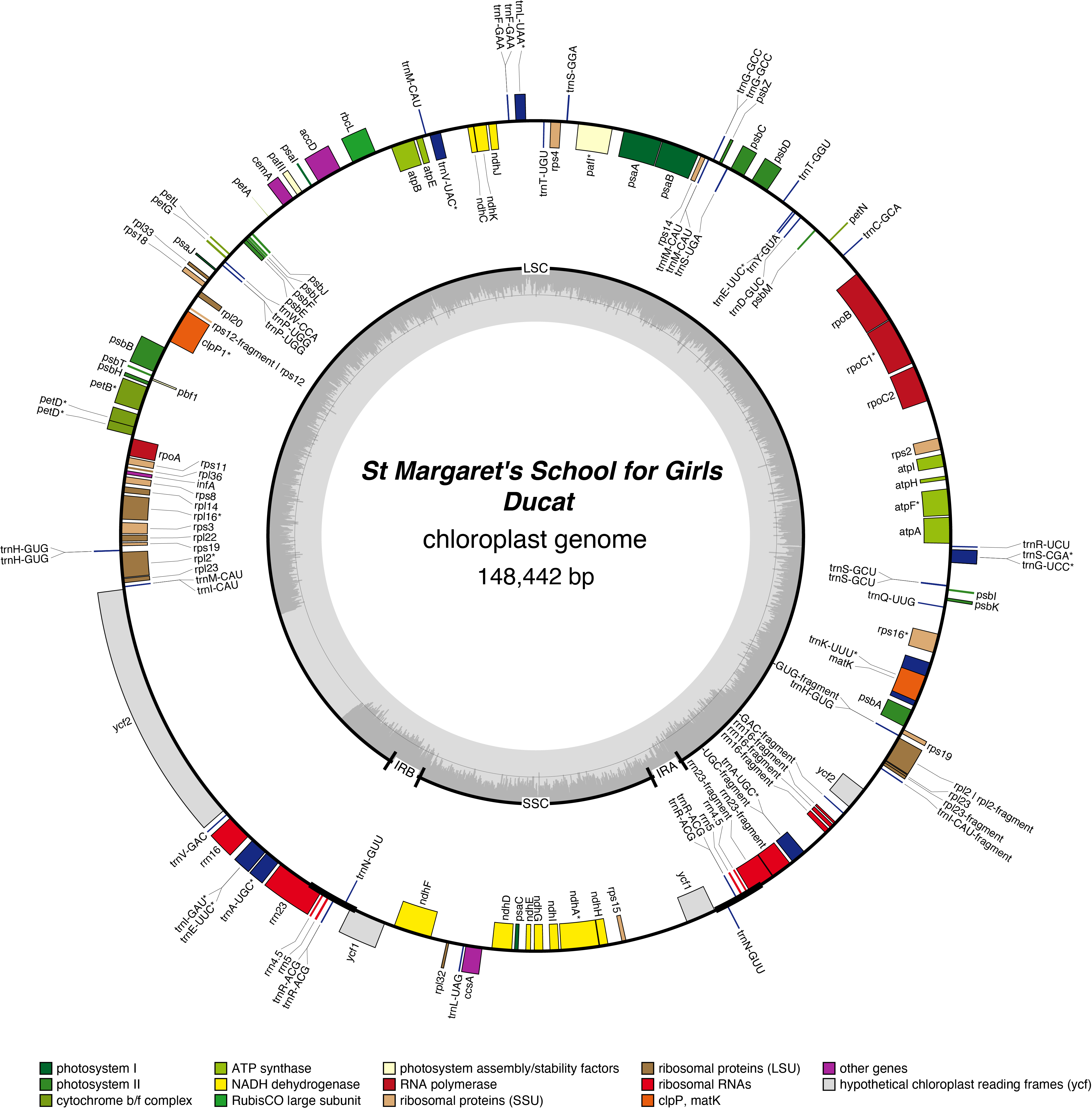
Ducat Chloroplast Genome
- Corona: Yellow
- Perianth: Yellow
- Division 1 (Trumpet Daffodils)
- Registered: <1907
| Total Sequenced Bases (bp)
help
|
18,496 | Mean Quality
help
|
5335.7 |
| Mean Read Length (bp)
help
|
14.0 | N50 Read Length (bp)
help
|
18.0 |
| Proportion of Chloroplast Bases (%)
help
|
0 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Loch Fyne Chloroplast Genome
- Corona: yellow
- Perianth: white
- Division: 2 (Large-Cupped Daffodils)
- Hybridiser: The Brodie of Brodie
- Registered: <1911

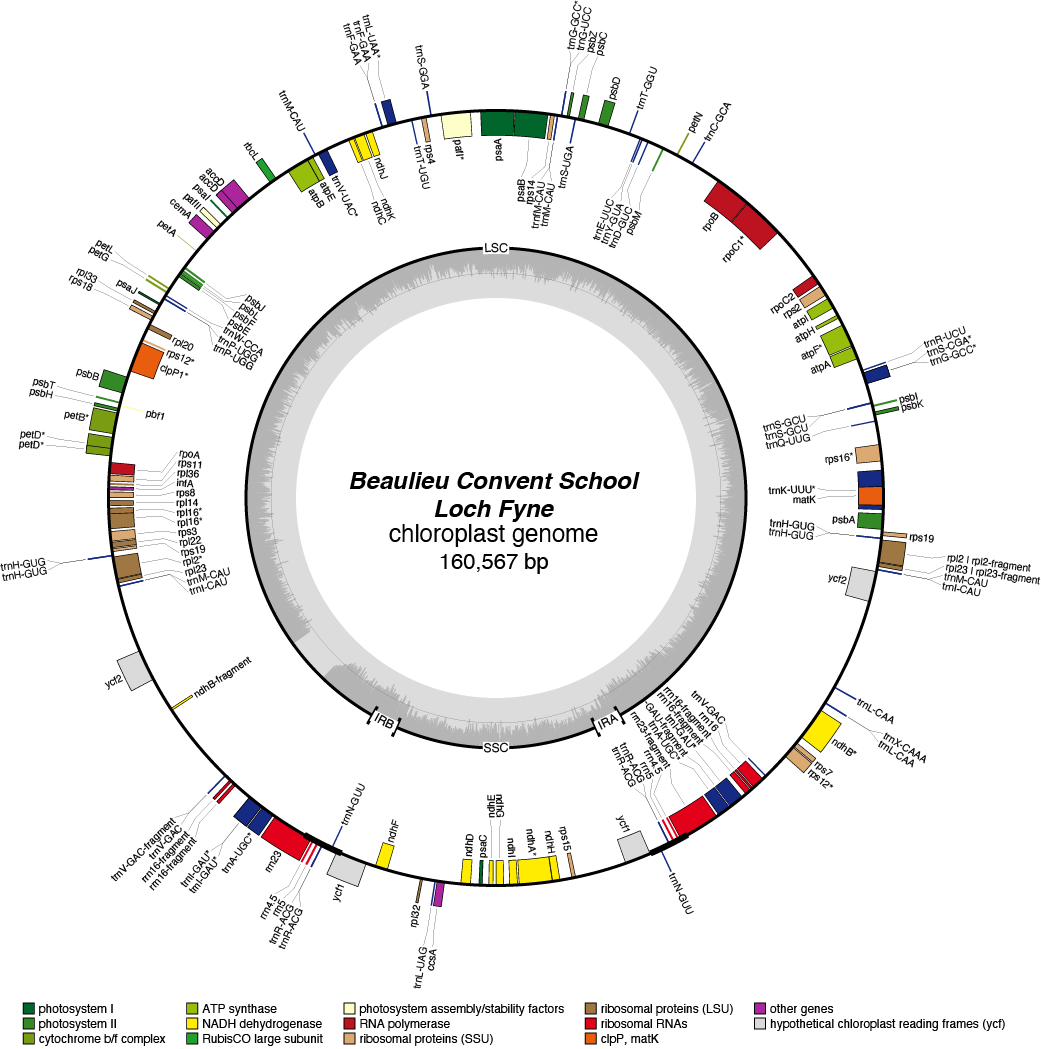
| Total Sequenced Bases (bp)
help
|
532,965,895 | Mean Quality
help
|
10.4 |
| Mean Read Length (bp)
help
|
1,851.7 | N50 Read Length (bp)
help
|
4,142.0 |
| Proportion of Chloroplast Bases (%)
help
|
4.7 | Chloroplast Coverage
help
|
156 |
| Assembled Contigs
help
|
6 | Assembled Contig Length (bp)
help
|
269177 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
160443 |
Pheasants Eye Chloroplast Genome
- Perianth: white
- Division 13 - Species, Wild Variants
| Total Sequenced Bases (bp)
help
|
1,022,814,204 | Mean Quality
help
|
11.5 |
| Mean Read Length (bp)
help
|
2,939.9 | N50 Read Length (bp)
help
|
7,052.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.7 | Chloroplast Coverage
help
|
105 |
| Assembled Contigs
help
|
4 | Assembled Contig Length (bp)
help
|
137850 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
262913 |
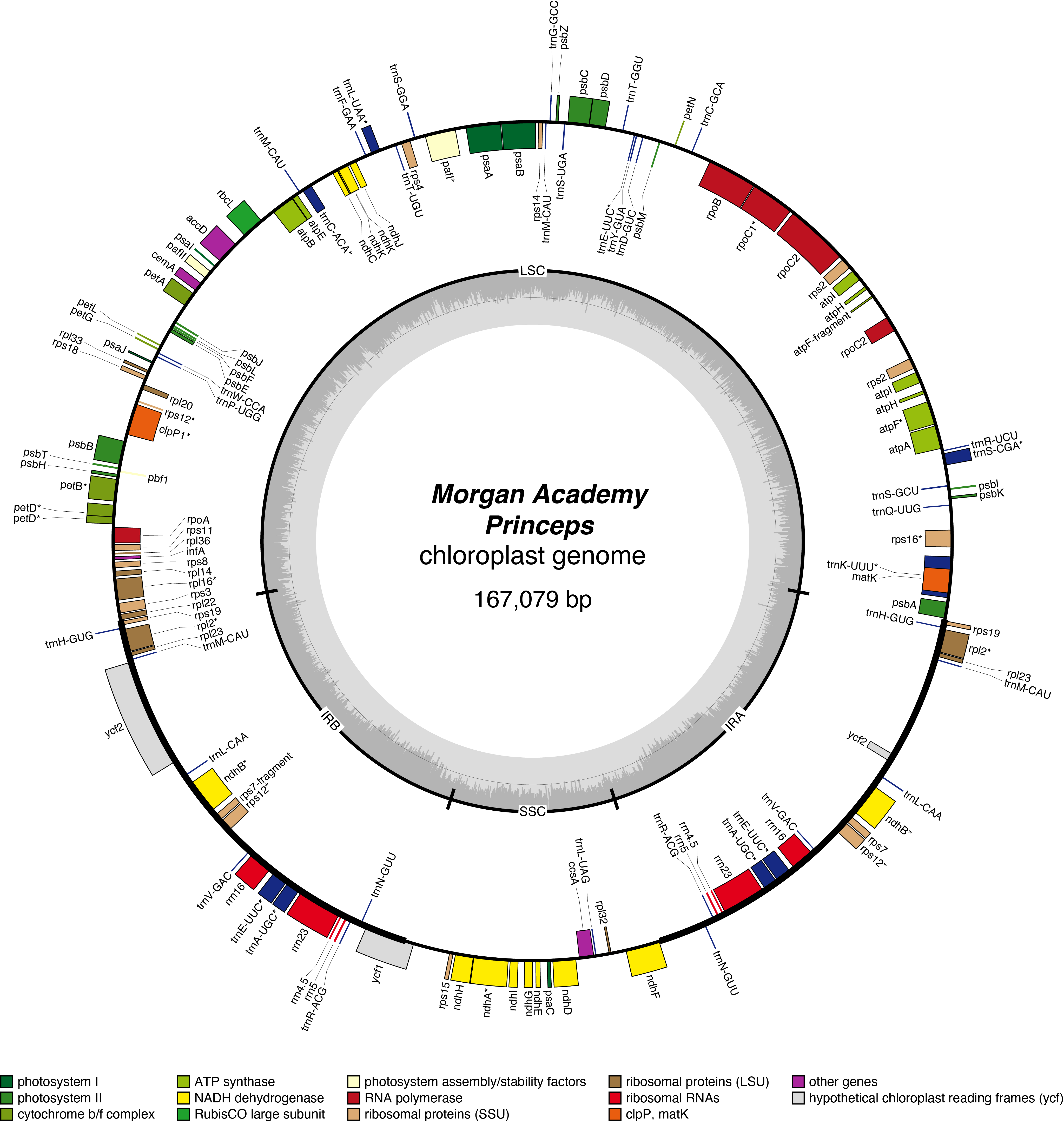
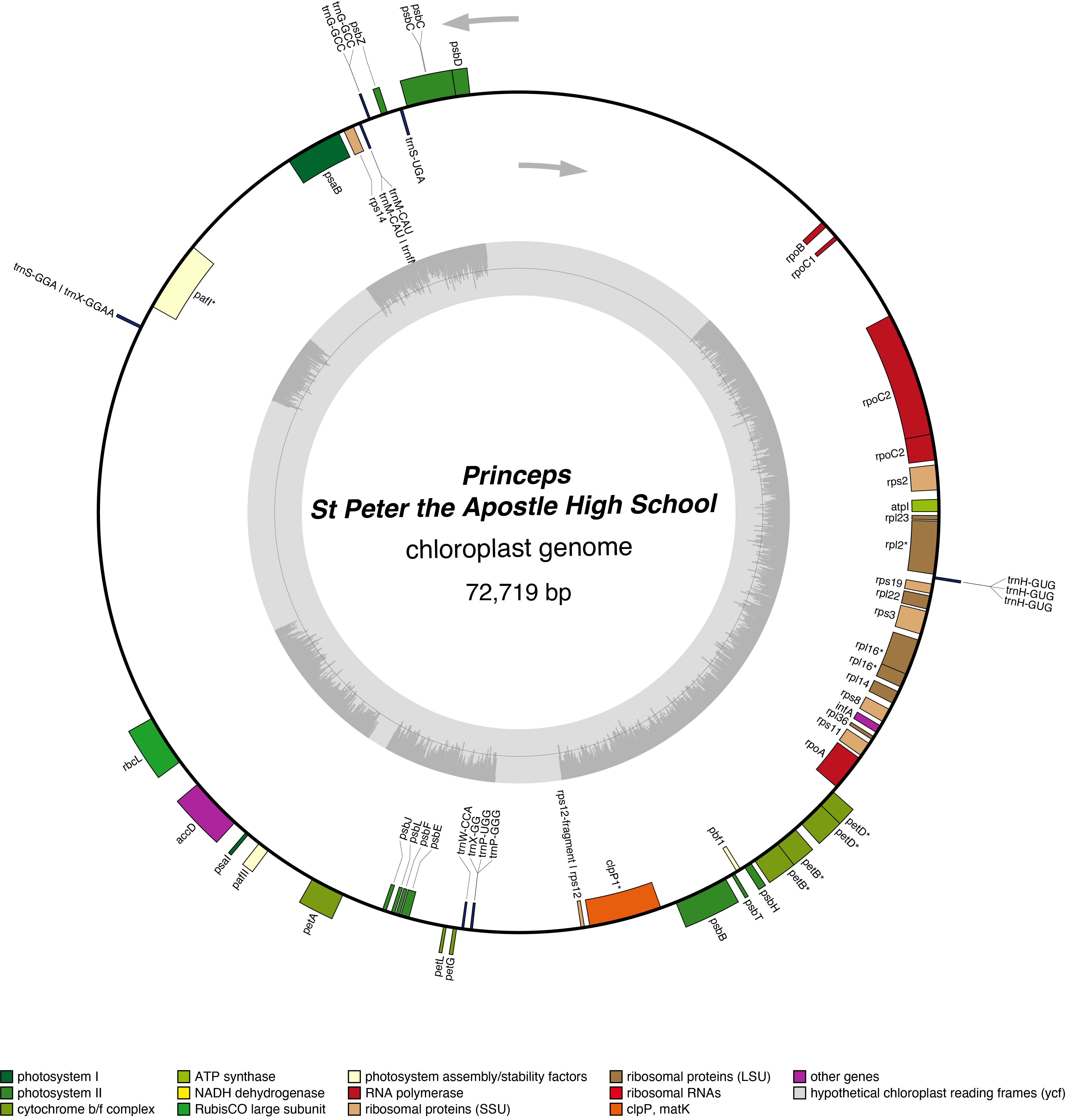
Princeps Chloroplast Genome
- Perianth: white
- Division 1 (Trumpet Daffodils)
- Hybridiser: Unknown
- Registered: <1830

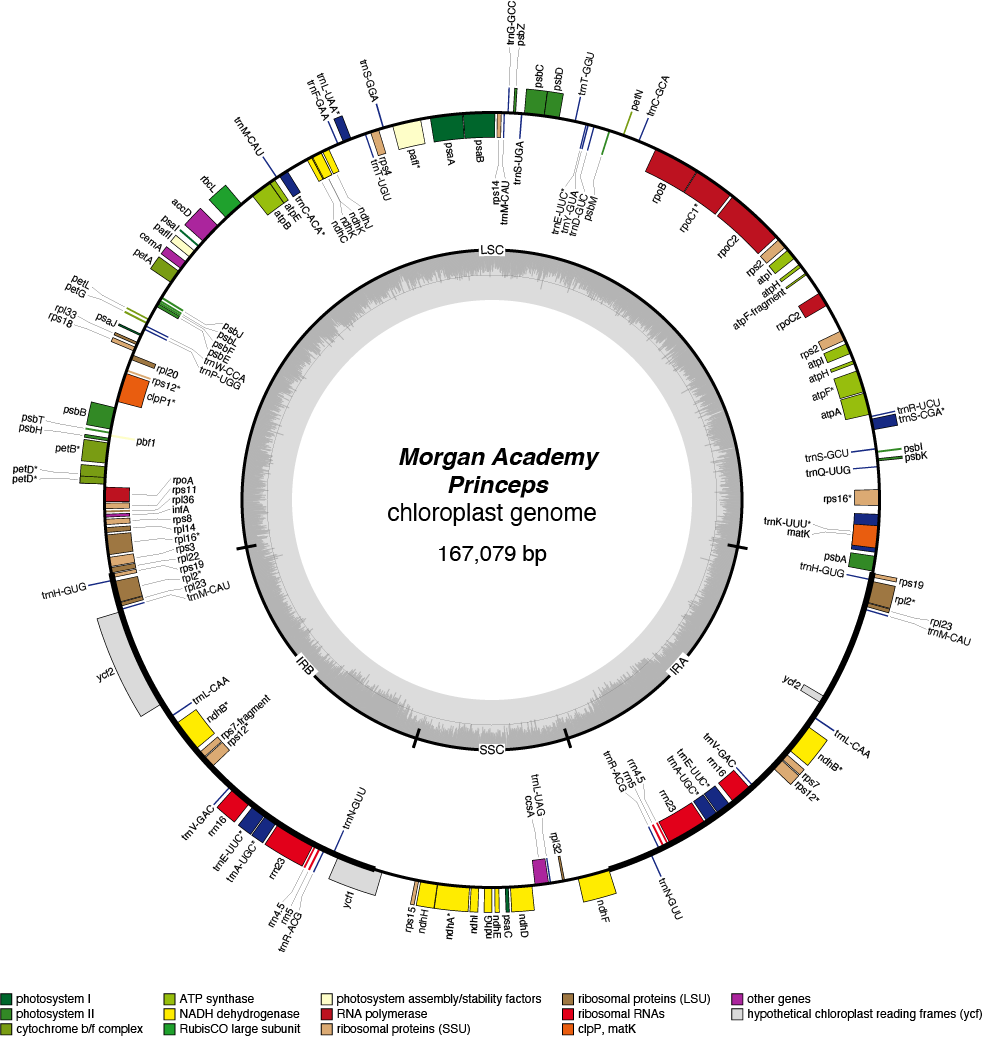
| Total Sequenced Bases (bp)
help
|
566,883,659 | Mean Quality
help
|
15.2 |
| Mean Read Length (bp)
help
|
4,825.4 | N50 Read Length (bp)
help
|
9,034.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.77 | Chloroplast Coverage
help
|
27 |
| Assembled Contigs
help
|
1 | Assembled Contig Length (bp)
help
|
167168 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
319111 |
Tahiti Chloroplast Genome
- Corona: orange
- Perianth: yellow
- Division: 4 (Double daffodils)
- Hybridiser: J.L. Richardson
- Registered: 1956
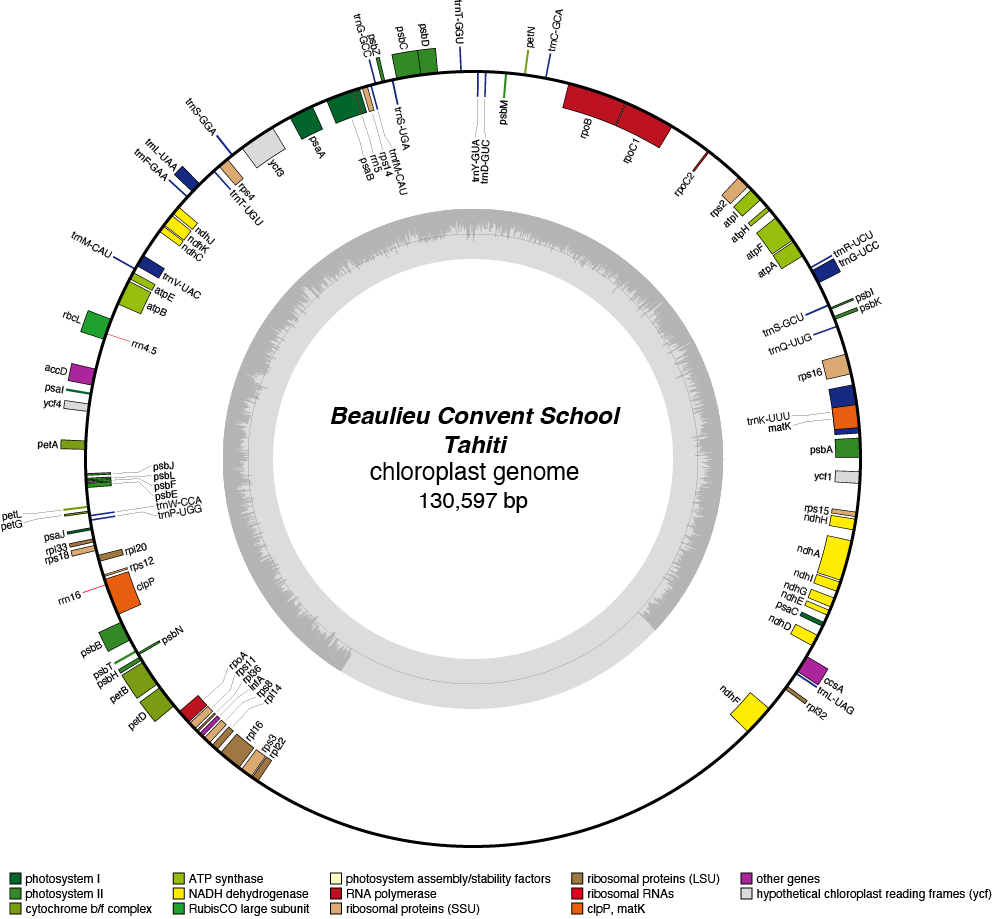
| Total Sequenced Bases (bp)
help
|
200,742,020 | Mean Quality
help
|
10.4 |
| Mean Read Length (bp)
help
|
2,893.1 | N50 Read Length (bp)
help
|
5,325.0 |
| Proportion of Chloroplast Bases (%)
help
|
4.8 | Chloroplast Coverage
help
|
59 |
| Assembled Contigs
help
|
5 | Assembled Contig Length (bp)
help
|
139689 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
130690 |
2023 Daffodils
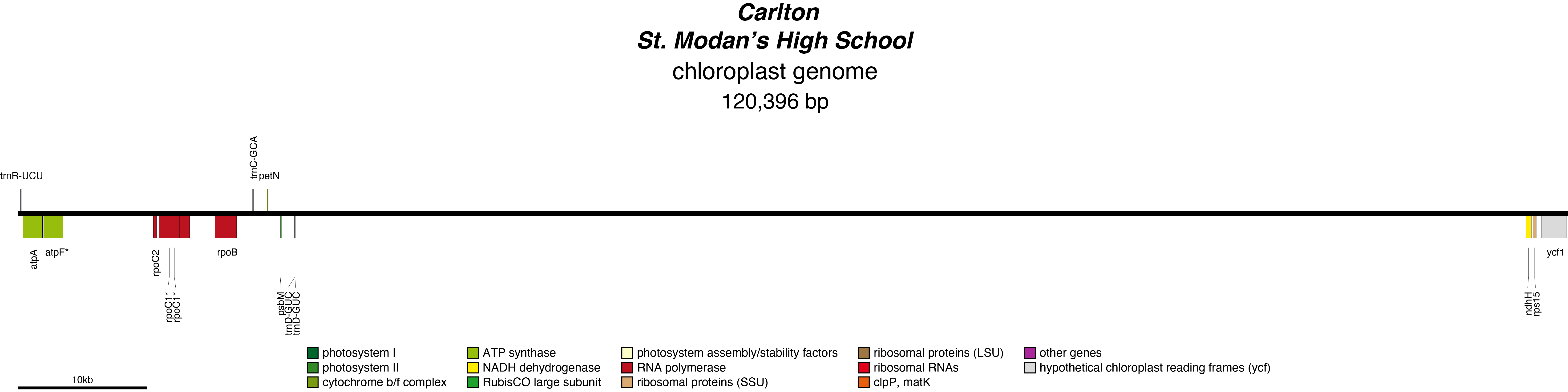
Carlton Chloroplast Genome
- Perianth: Yellow
- Corona: Yellow
- Division 2 (Large cupped daffodils)
- Hybridiser: P.D. Williams
- Registered: <1927
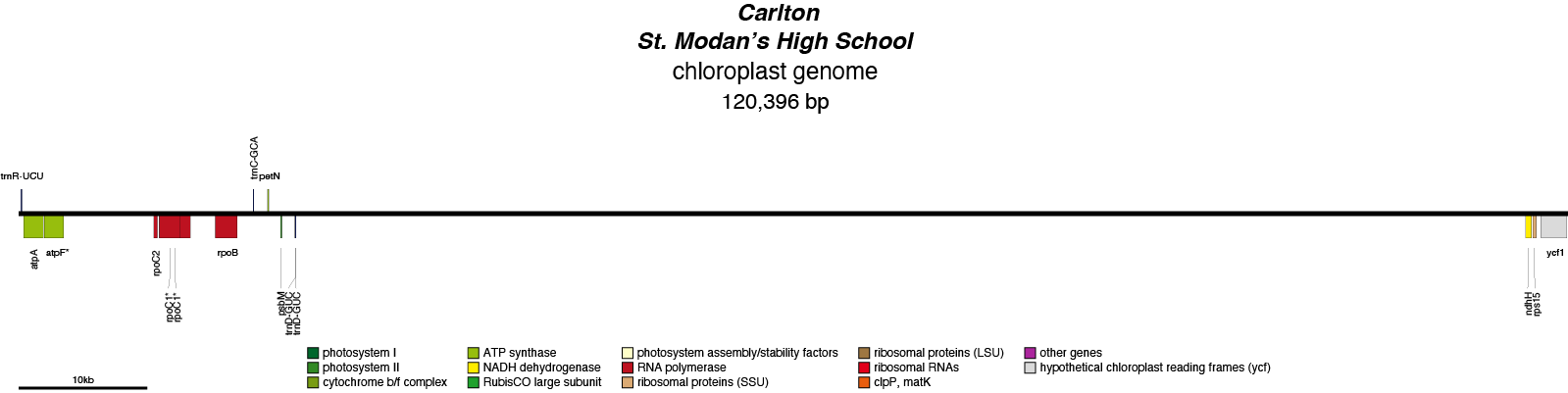
| Total Sequenced Bases (bp)
help
|
329,228,066 | Mean Quality
help
|
10.3 |
| Mean Read Length (bp)
help
|
470.6 | N50 Read Length (bp)
help
|
525.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.62 | Chloroplast Coverage
help
|
12 |
| Assembled Contigs
help
|
5 | Assembled Contig Length (bp)
help
|
19047 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
120354 |
Castermoon Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
104,398,663 | Mean Quality
help
|
15.4 |
| Mean Read Length (bp)
help
|
452.2 | N50 Read Length (bp)
help
|
571.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.7 | Chloroplast Coverage
help
|
11 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Copper Bowl Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
492,962,811 | Mean Quality
help
|
15.4 |
| Mean Read Length (bp)
help
|
441.6 | N50 Read Length (bp)
help
|
474.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.24 | Chloroplast Coverage
help
|
7 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Coulmony Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
630,205,582 | Mean Quality
help
|
15.6 |
| Mean Read Length (bp)
help
|
470.9 | N50 Read Length (bp)
help
|
517.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.62 | Chloroplast Coverage
help
|
24 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
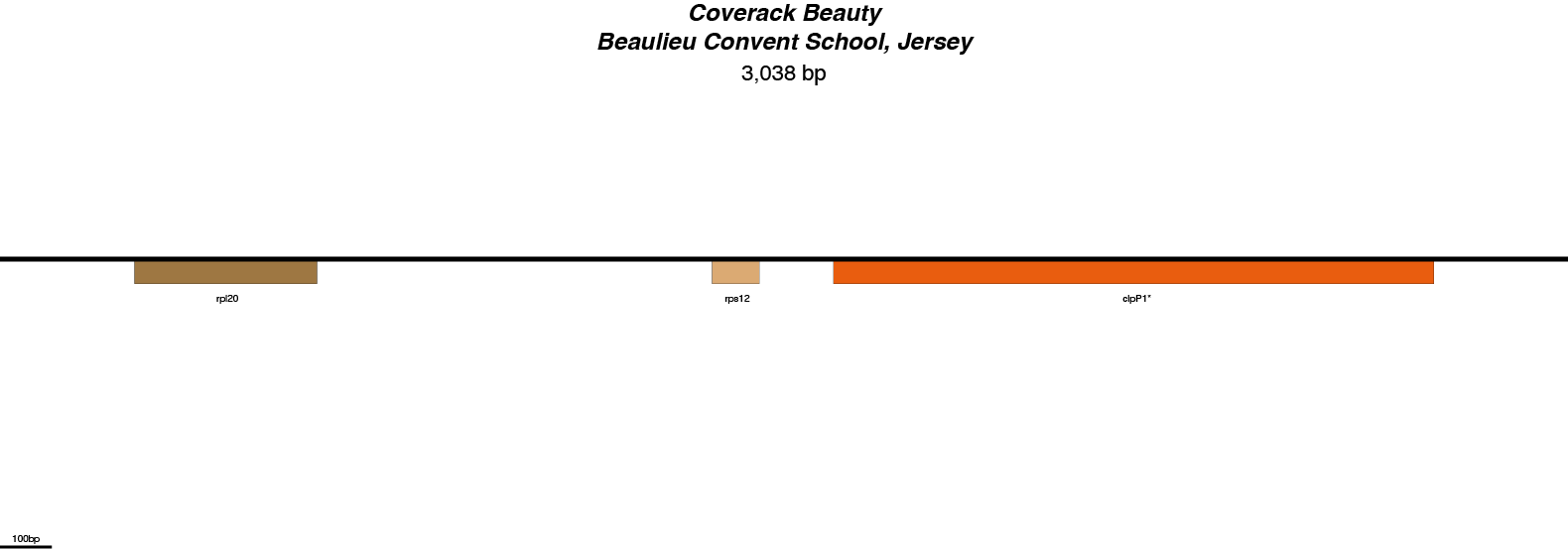
Coverack Beauty Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
657,204,933 | Mean Quality
help
|
15.8 |
| Mean Read Length (bp)
help
|
537.9 | N50 Read Length (bp)
help
|
638.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.9 | Chloroplast Coverage
help
|
79 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
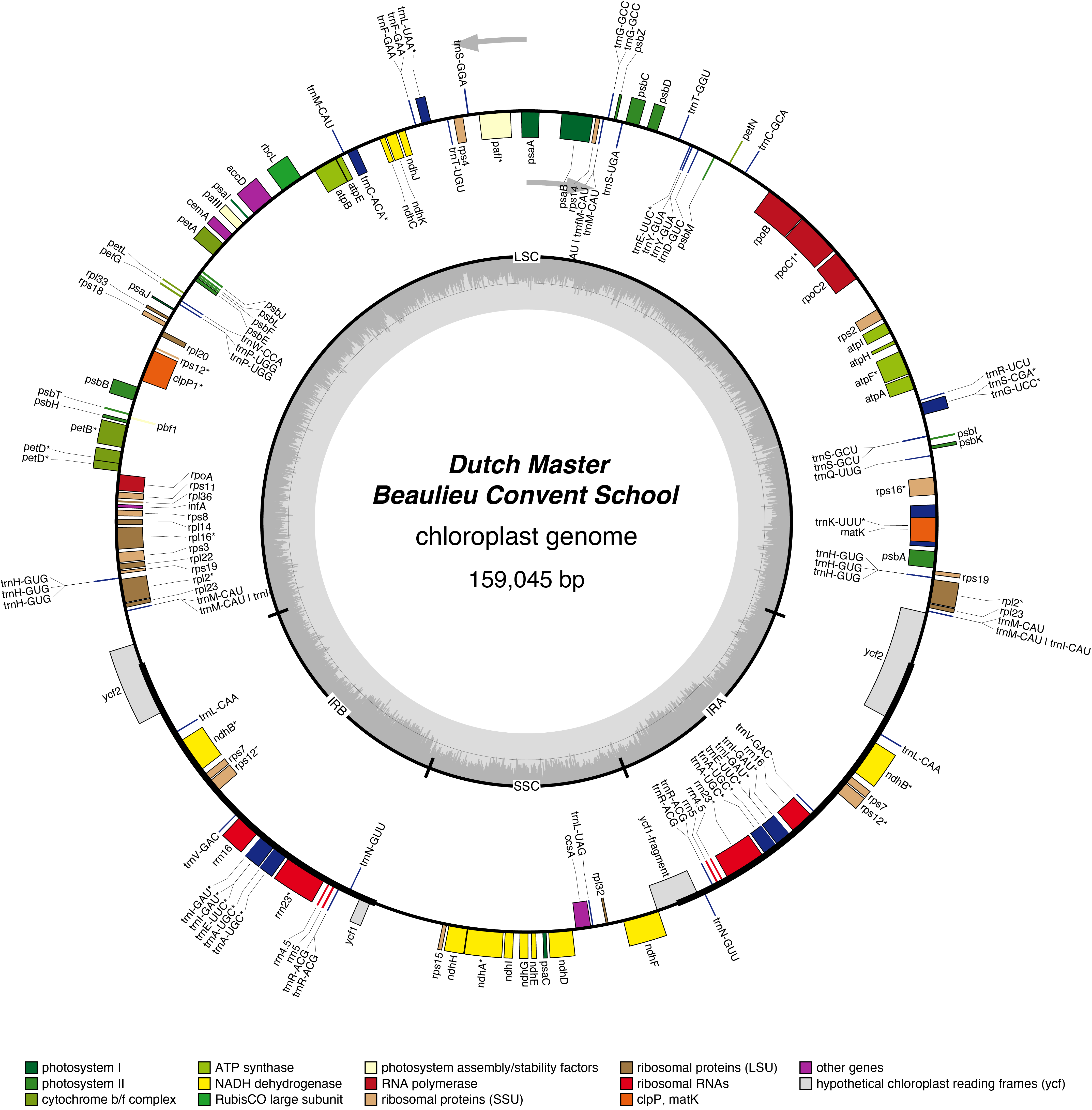
Dutch Master Chloroplast Genome
- Perianth: yellow
- Corona: yellow
- Division 7 (Jonquilla Daffodils)
- Hybridiser: Matthew Zandbergen
- Registered: 1948

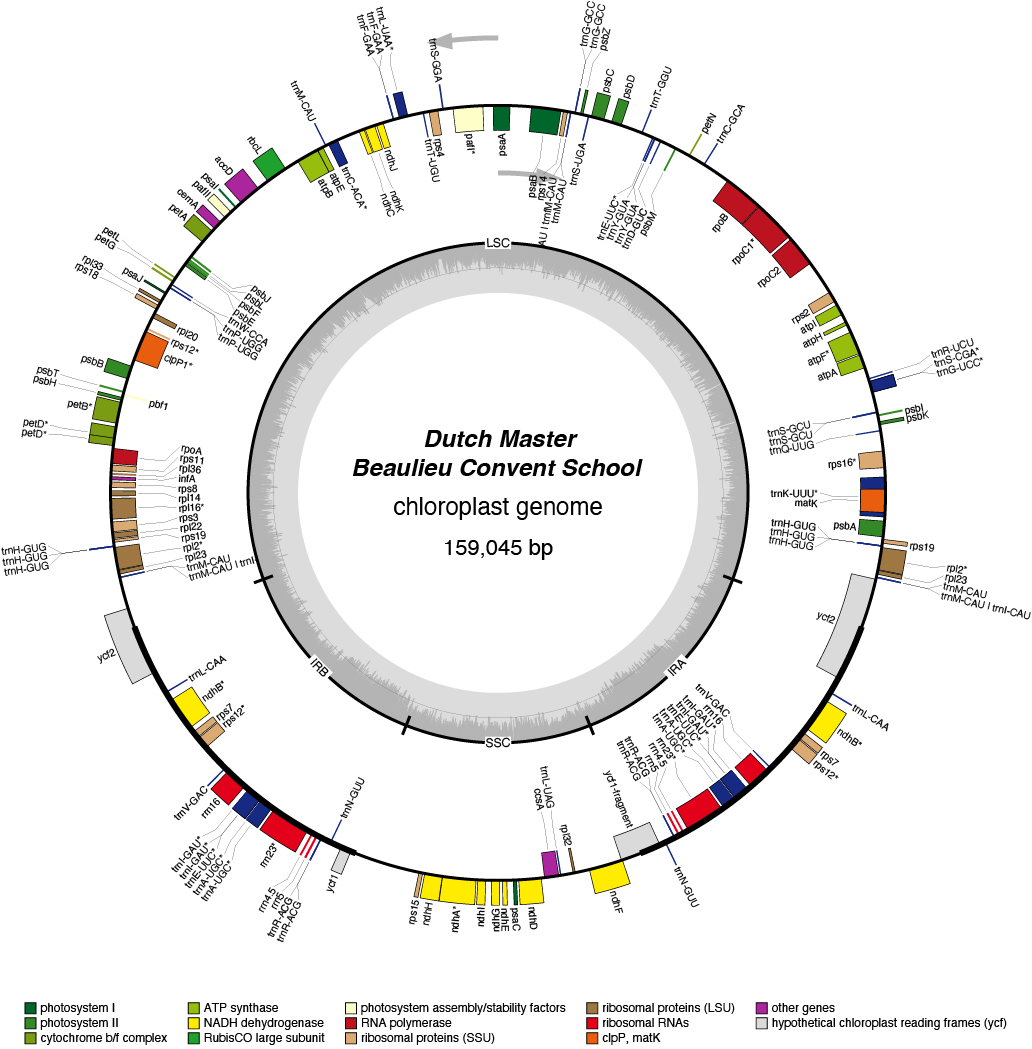
| Total Sequenced Bases (bp)
help
|
778,191,977 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
3,036.5 | N50 Read Length (bp)
help
|
5,395.0 |
| Proportion of Chloroplast Bases (%)
help
|
4.5 | Chloroplast Coverage
help
|
217 |
| Assembled Contigs
help
|
1 | Assembled Contig Length (bp)
help
|
184643 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
159064 |
Ferness Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
579,724,008 | Mean Quality
help
|
15.6 |
| Mean Read Length (bp)
help
|
420.1 | N50 Read Length (bp)
help
|
439.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.14 | Chloroplast Coverage
help
|
4 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
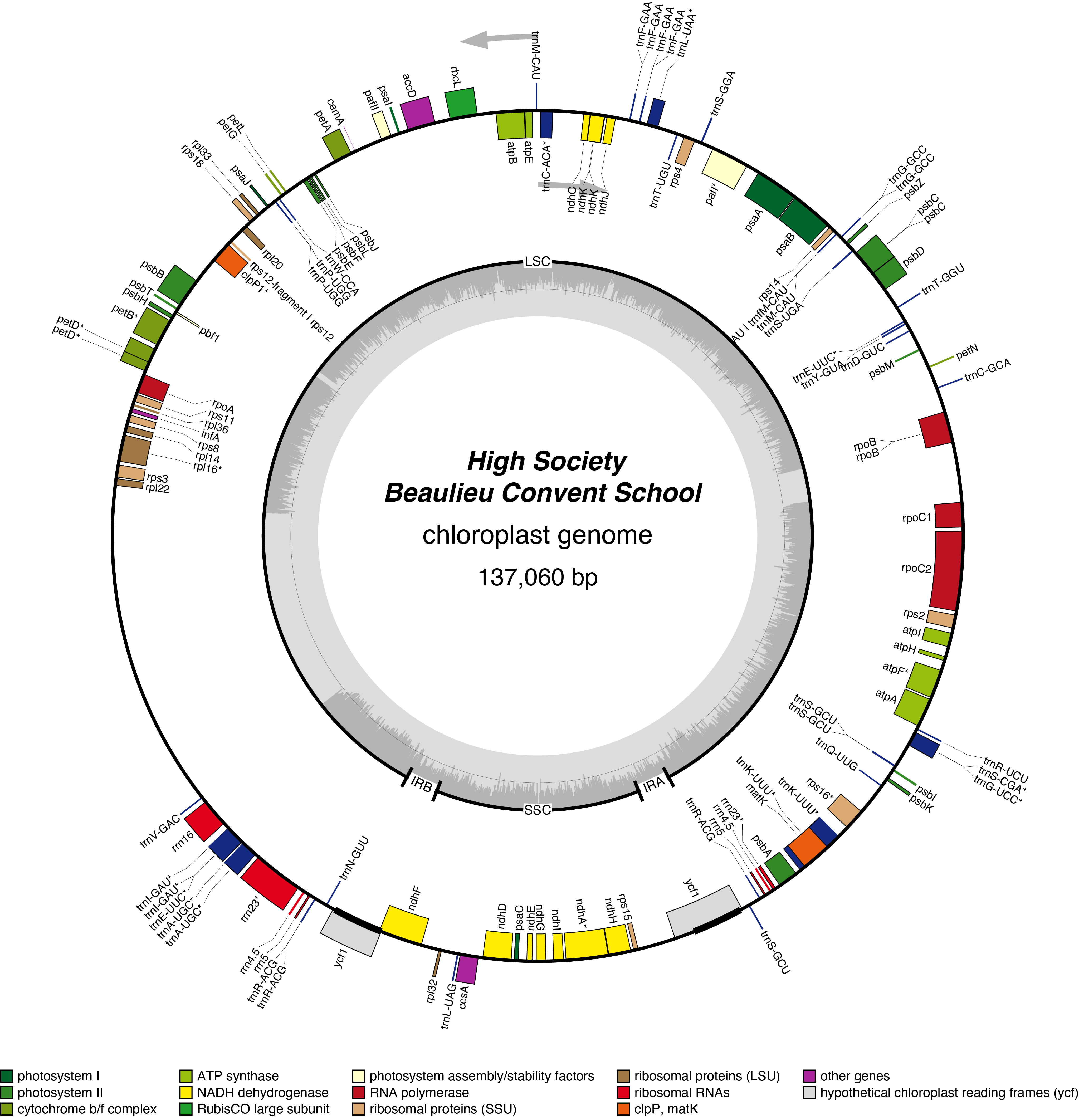
High Society Chloroplast Genome
- Perianth: white
- Corona: green/white/pink
- Pollen parent: Richardson
- Seed parent: May Queen
- Division 2 (Large cupped daffodils)
- Hybridiser: Brian Duncan
- Registered: 1979
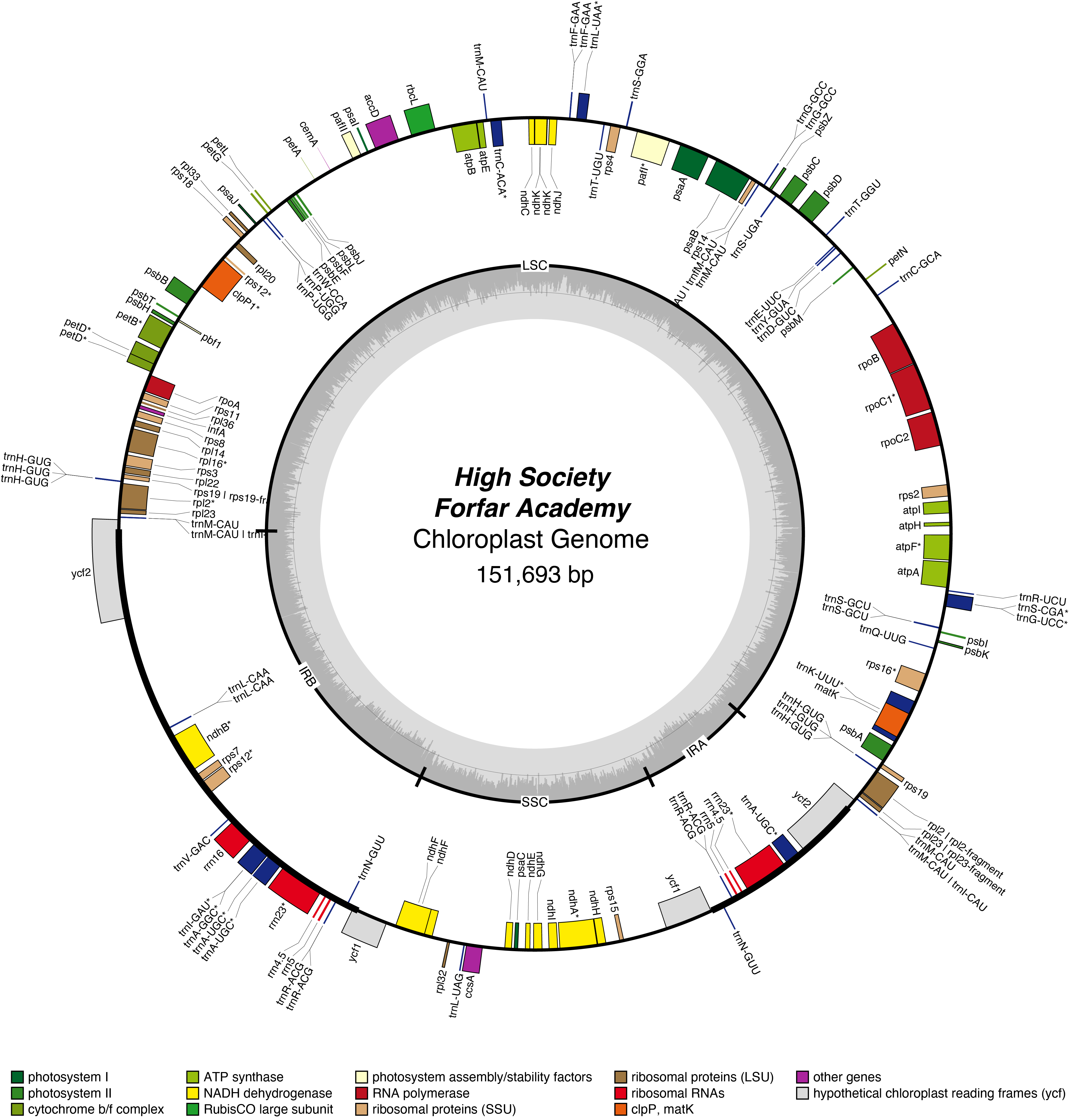
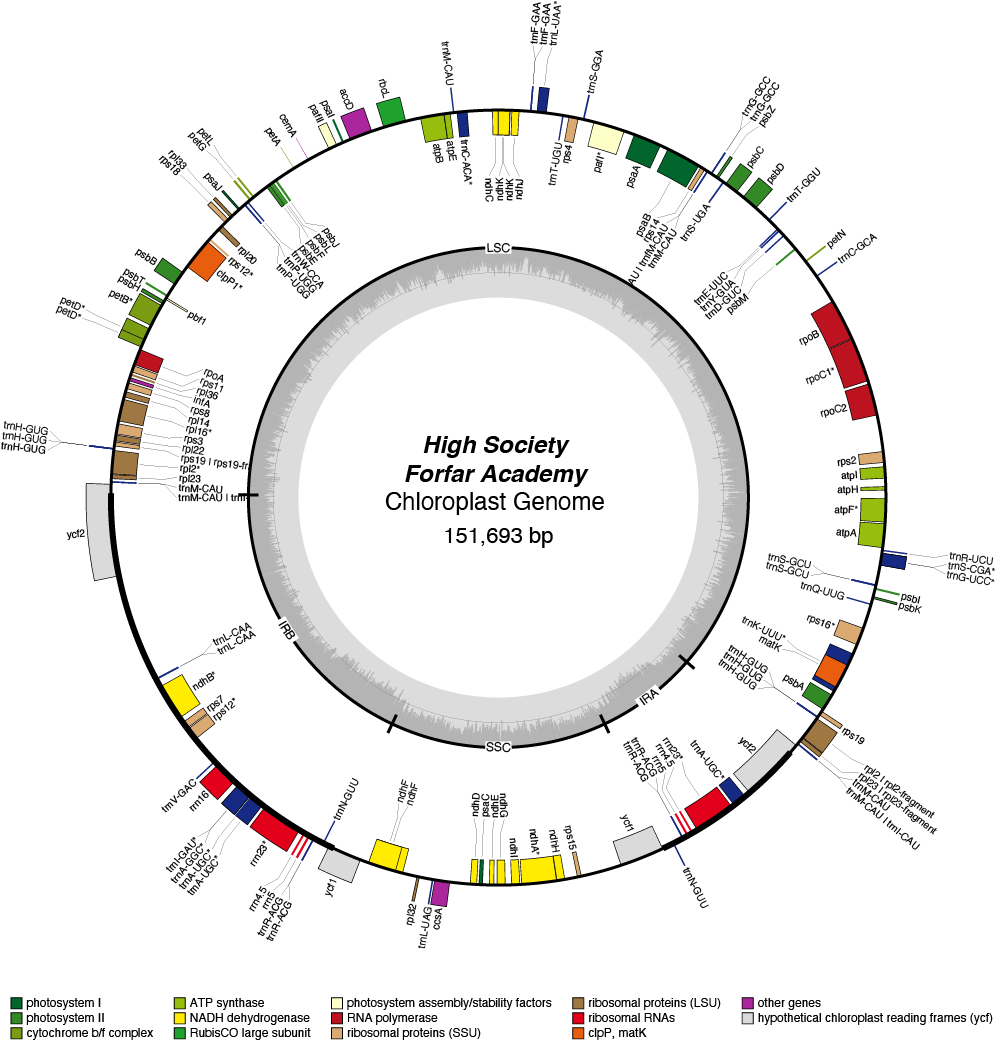
| Total Sequenced Bases (bp)
help
|
2,088,574,589 | Mean Quality
help
|
10.6 |
| Mean Read Length (bp)
help
|
1,455.0 | N50 Read Length (bp)
help
|
2,173.0 |
| Proportion of Chloroplast Bases (%)
help
|
2.3 | Chloroplast Coverage
help
|
298 |
| Assembled Contigs
help
|
3029 | Assembled Contig Length (bp)
help
|
21321774 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
151524 |
High Society Chloroplast Genome
- Perianth: white
- Corona: green/white/pink
- Pollen parent: Richardson
- Seed parent: May Queen
- Division 2 (Large cupped daffodils)
- Hybridiser: Brian Duncan
- Registered: 1979

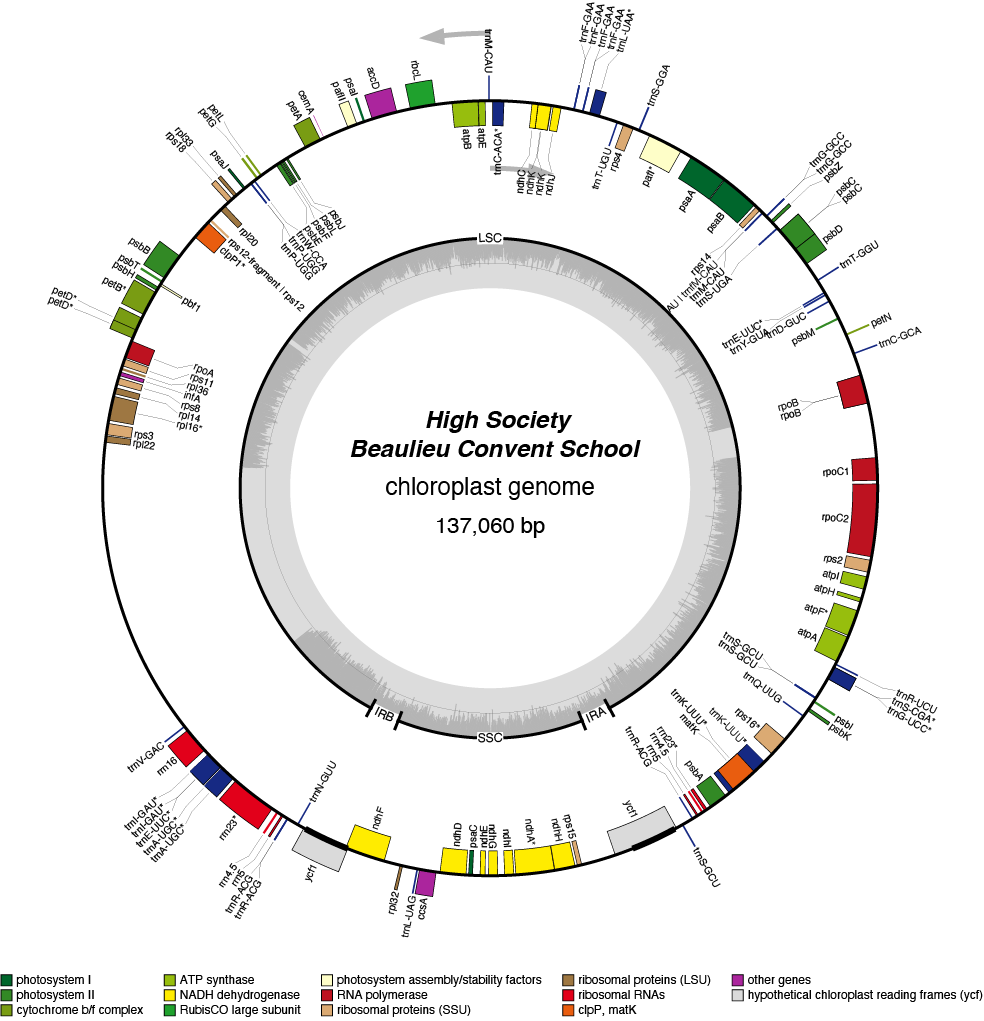
| Total Sequenced Bases (bp)
help
|
193,339,657 | Mean Quality
help
|
10.4 |
| Mean Read Length (bp)
help
|
2,434.5 | N50 Read Length (bp)
help
|
4,564.0 |
| Proportion of Chloroplast Bases (%)
help
|
2.5 | Chloroplast Coverage
help
|
30 |
| Assembled Contigs
help
|
7 | Assembled Contig Length (bp)
help
|
117365 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
137260 |
Lowin Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
189,249,161 | Mean Quality
help
|
14.9 |
| Mean Read Length (bp)
help
|
307.6 | N50 Read Length (bp)
help
|
363.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.47 | Chloroplast Coverage
help
|
5 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
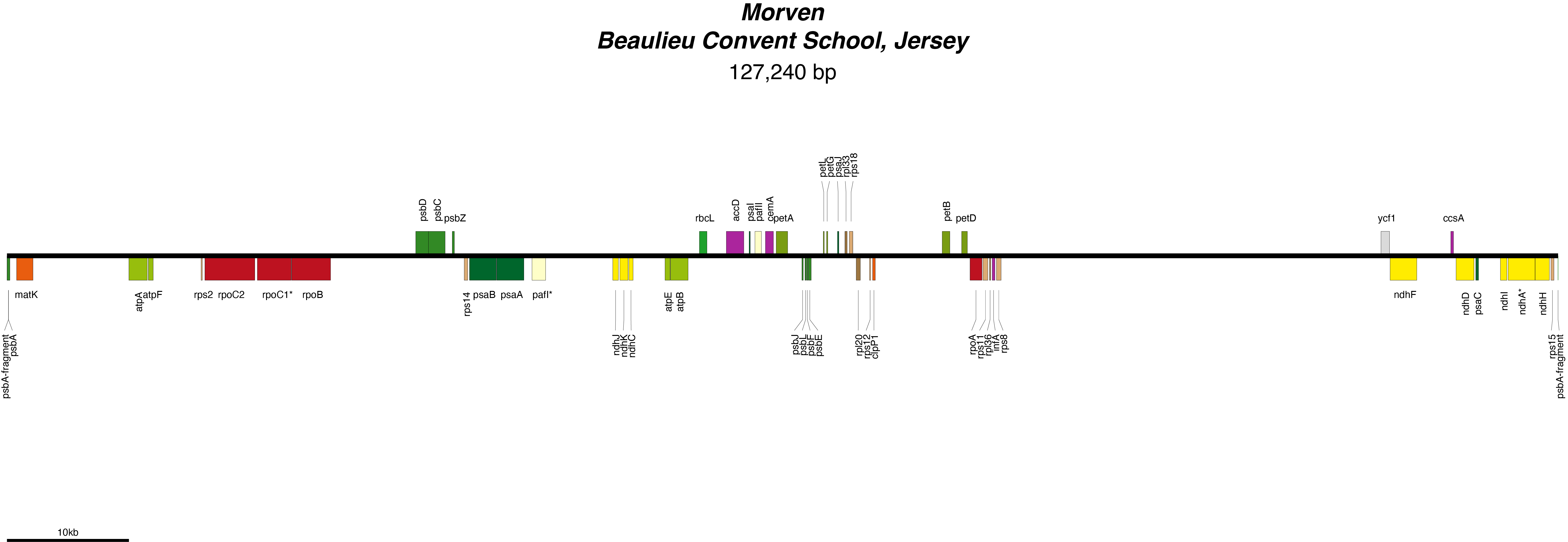
Morven Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
29,808,208 | Mean Quality
help
|
14.6 |
| Mean Read Length (bp)
help
|
238.4 | N50 Read Length (bp)
help
|
268.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.012 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Princeps Chloroplast Genome
- Perianth: white
- Division 1 (Trumpet Daffodils)
- Hybridiser: Unknown
- Registered: <1830
| Total Sequenced Bases (bp)
help
|
71,513,017 | Mean Quality
help
|
10.4 |
| Mean Read Length (bp)
help
|
655.3 | N50 Read Length (bp)
help
|
819.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.57 | Chloroplast Coverage
help
|
2 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
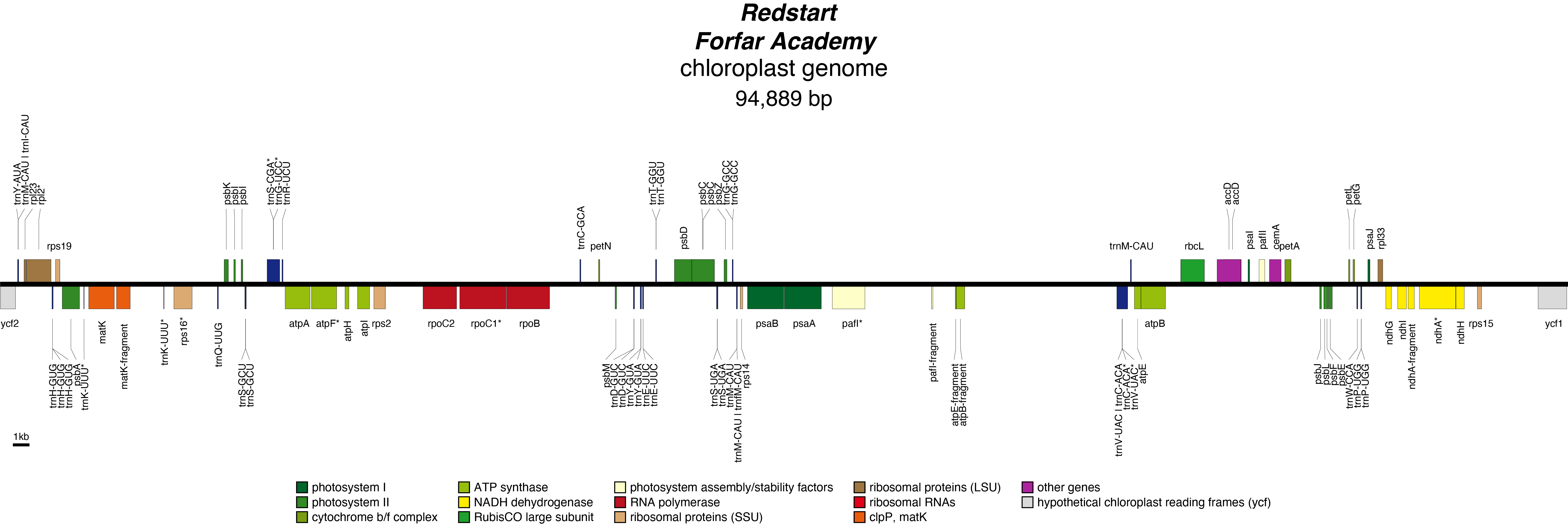
Redstart Chloroplast Genome
Thanks to the Hardy Plant Society for the image.
- Perianth: white
- Corona: White with a green base and orange rim
- Pollen parent: Sylvia O'Neill
- Seed parent: Rubra
- Division 3 (Small cupped daffodils)
- Hybridiser: G.E.Mitsch
- Registered: 1959

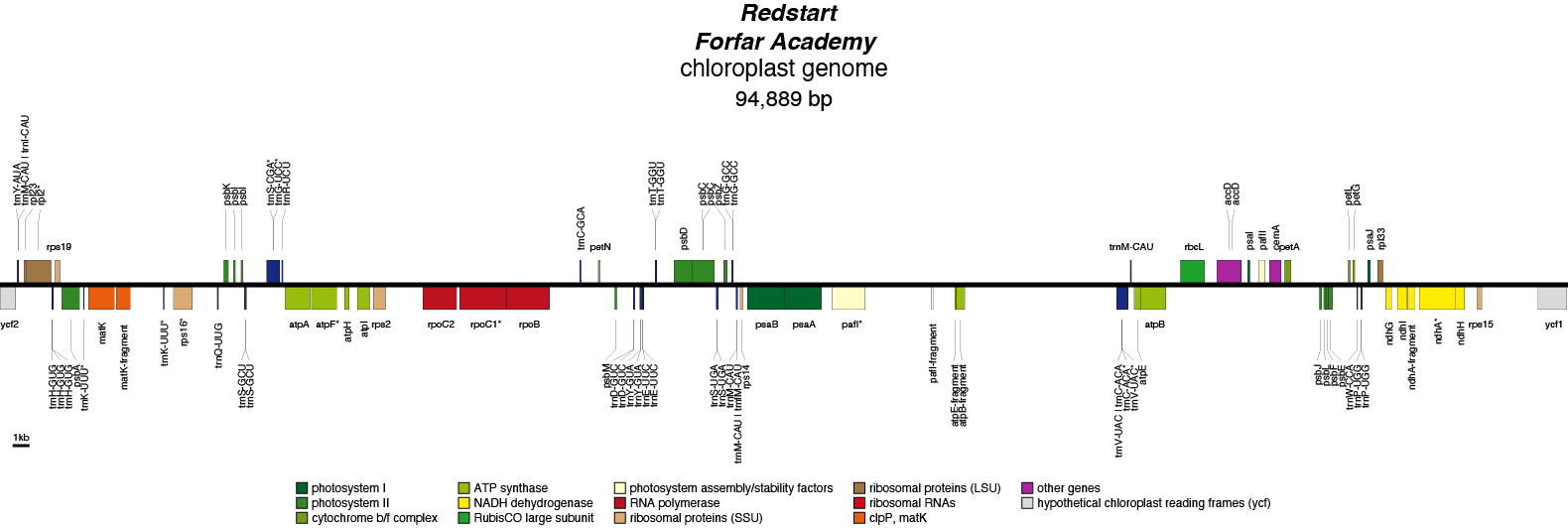
| Total Sequenced Bases (bp)
help
|
637,075,774 | Mean Quality
help
|
10.4 |
| Mean Read Length (bp)
help
|
974.3 | N50 Read Length (bp)
help
|
1,280.0 |
| Proportion of Chloroplast Bases (%)
help
|
2.8 | Chloroplast Coverage
help
|
110 |
| Assembled Contigs
help
|
21 | Assembled Contig Length (bp)
help
|
157859 |
| Assembled Scaffolds
help
|
2 | Assembled Scaffold Length (bp)
help
|
159759 |
Rose of Cuan Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
391,793,982 | Mean Quality
help
|
15.6 |
| Mean Read Length (bp)
help
|
417.7 | N50 Read Length (bp)
help
|
465.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.45 | Chloroplast Coverage
help
|
10 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
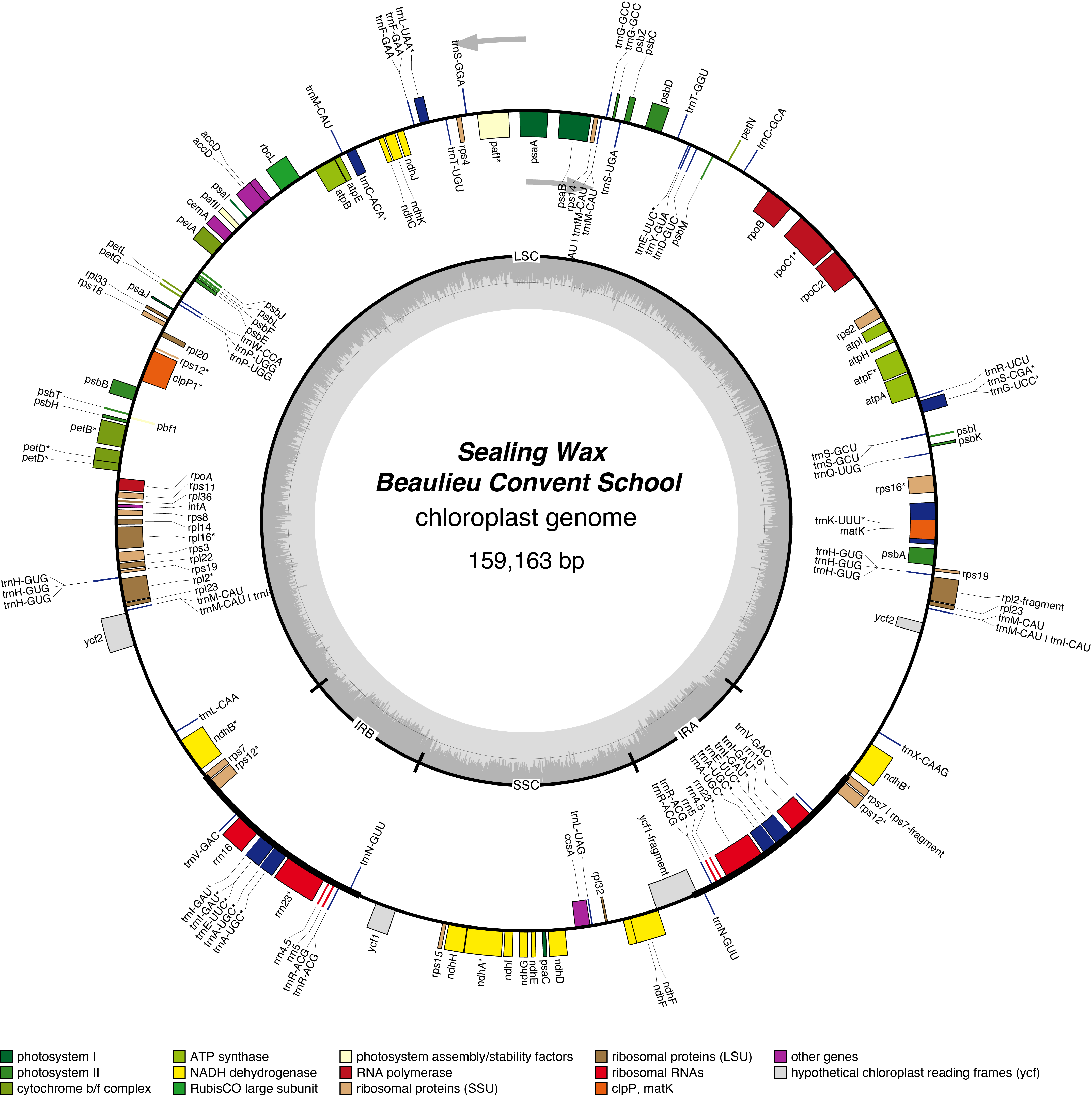
Sealing Wax Chloroplast Genome
- Perianth: yellow
- Corona: orange
- Division 2 (Small cupped Daffodils)
- Hybridiser: Barr and Sons
- Registered: 1957

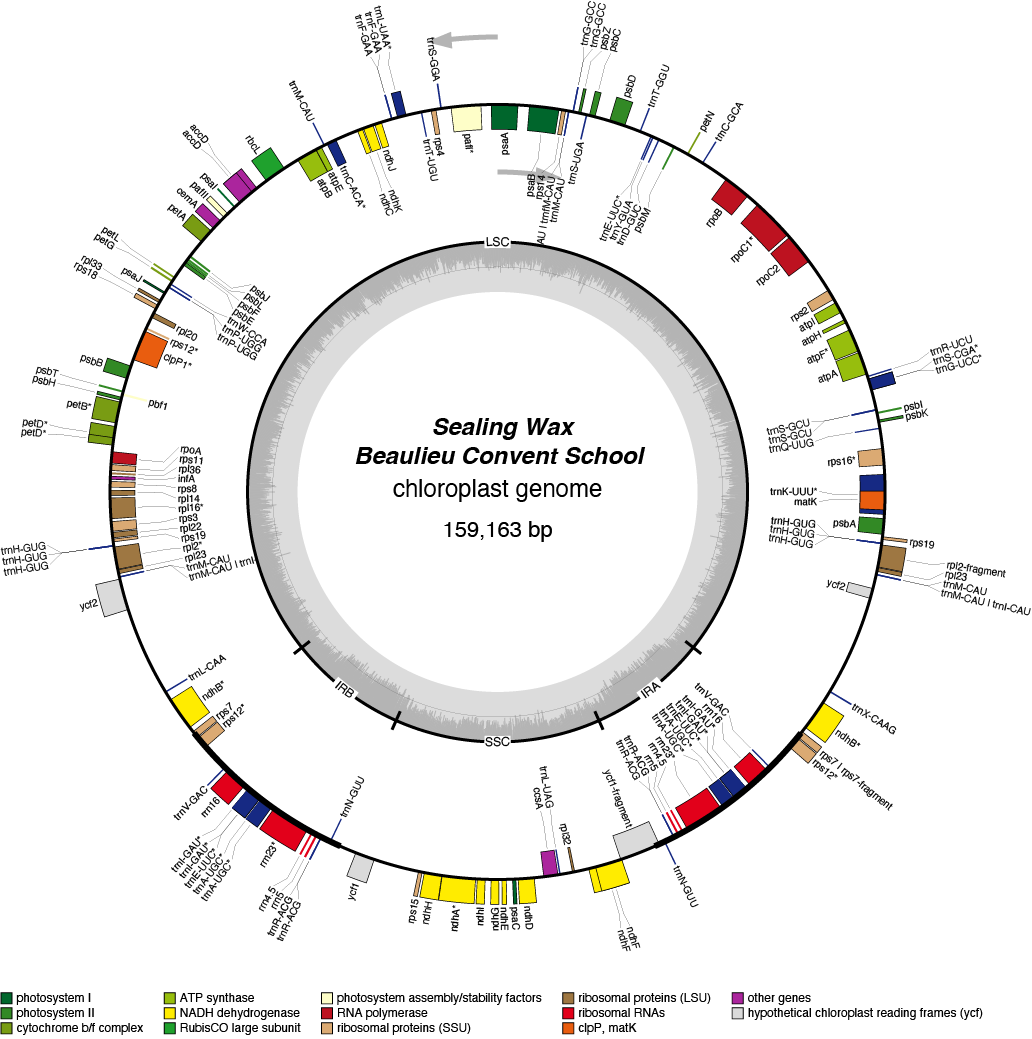
| Total Sequenced Bases (bp)
help
|
853,626,090 | Mean Quality
help
|
10.6 |
| Mean Read Length (bp)
help
|
858.2 | N50 Read Length (bp)
help
|
1,138.0 |
| Proportion of Chloroplast Bases (%)
help
|
10 | Chloroplast Coverage
help
|
534 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
172528 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
159291 |
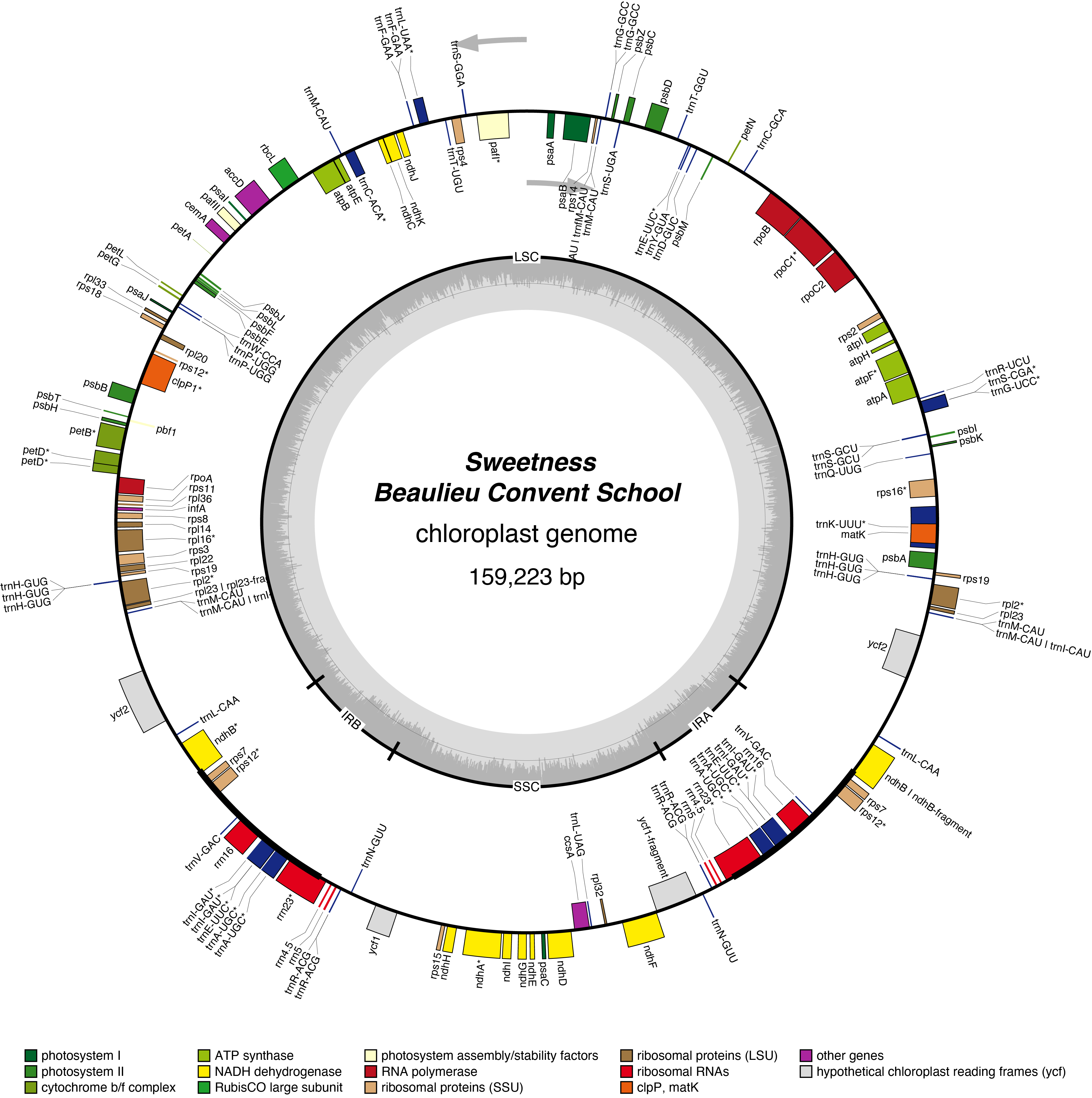
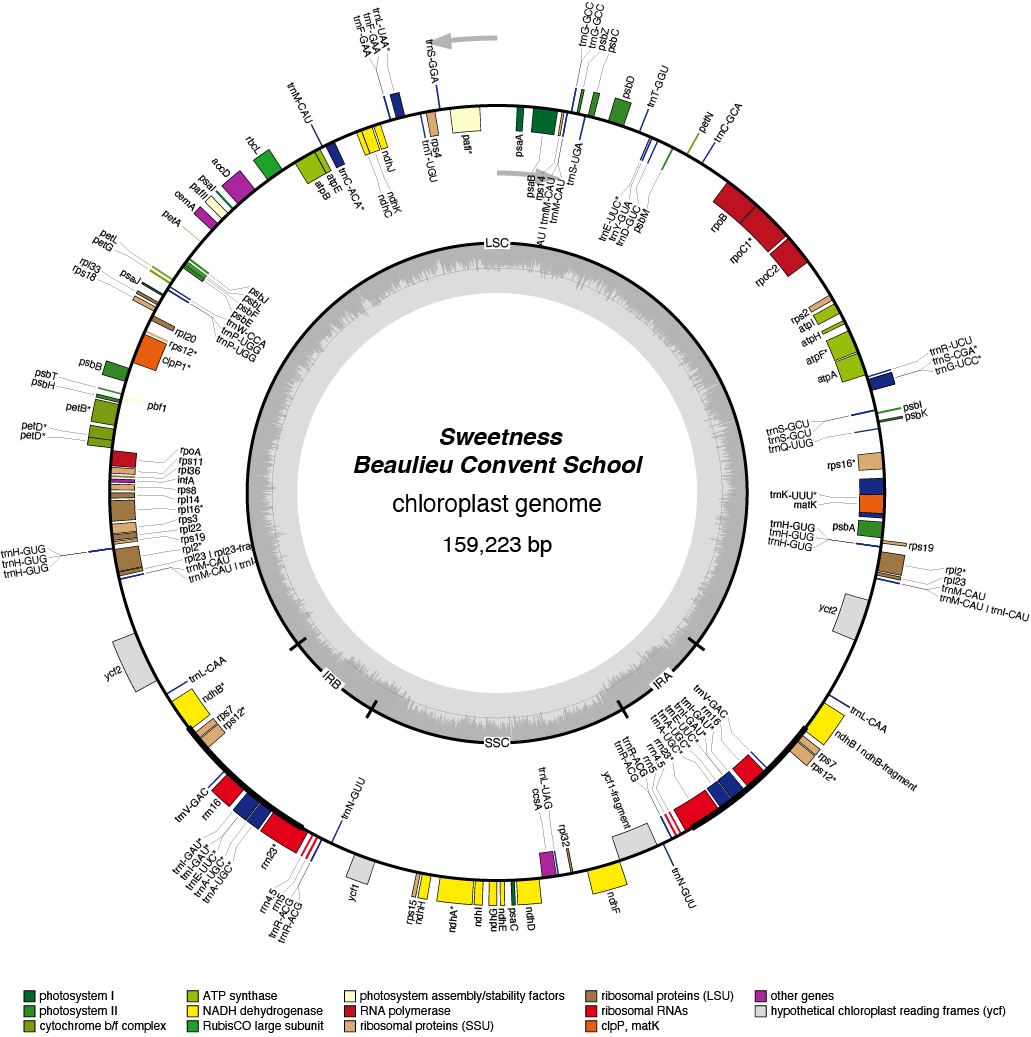
Sweetness Chloroplast Genome
- Perianth: yellow
- Corona: yellow
- Pollen parent: N. jonquilla
- Division 7 (Jonquilla Daffodils)
- Hybridiser: R.V. Favell
- Registered: <1939

| Total Sequenced Bases (bp)
help
|
117,375,135 | Mean Quality
help
|
10.1 |
| Mean Read Length (bp)
help
|
2,650.9 | N50 Read Length (bp)
help
|
4,438.0 |
| Proportion of Chloroplast Bases (%)
help
|
5.4 | Chloroplast Coverage
help
|
39 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
162909 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
159334 |
Therapia Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
774,466,195 | Mean Quality
help
|
15.7 |
| Mean Read Length (bp)
help
|
476.3 | N50 Read Length (bp)
help
|
501.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.16 | Chloroplast Coverage
help
|
7 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
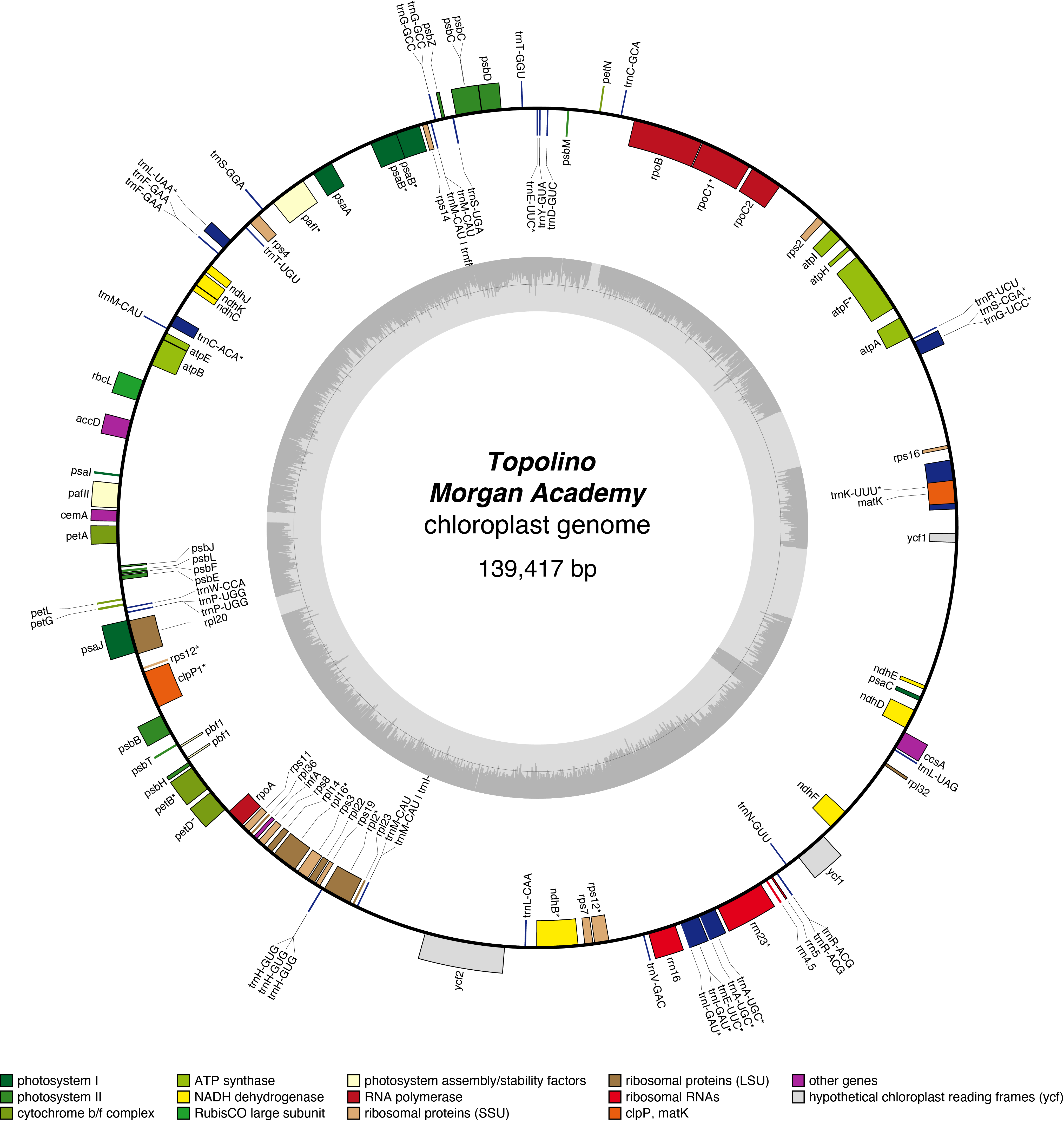
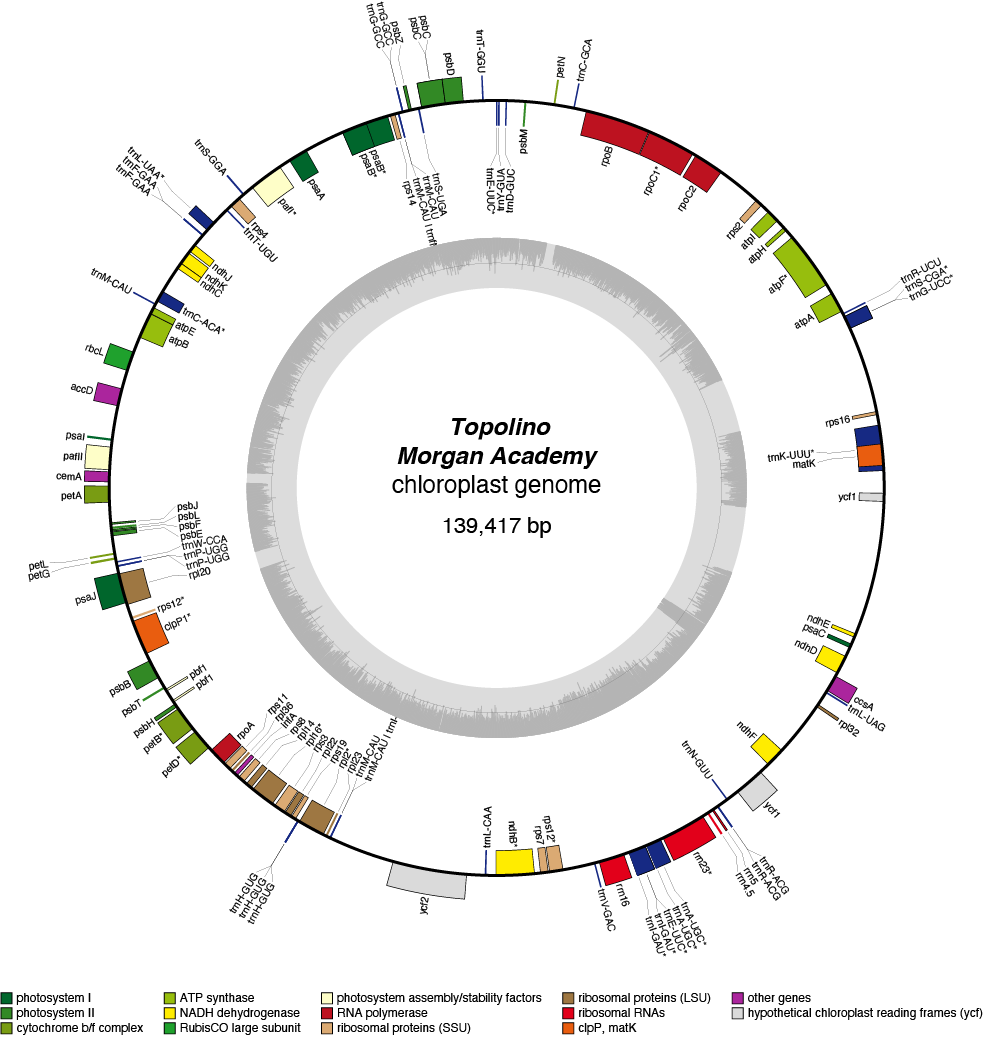
Topolino Chloroplast Genome
- Perianth: White
- Corona: yellow
- Division 1 (Trumpet daffodils)
- Hybridiser: J. Gerritsen & Son
- Registered: 1965
| Total Sequenced Bases (bp)
help
|
329,245,778 | Mean Quality
help
|
10.1 |
| Mean Read Length (bp)
help
|
1,028.3 | N50 Read Length (bp)
help
|
1,552.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.82 | Chloroplast Coverage
help
|
16 |
| Assembled Contigs
help
|
427 | Assembled Contig Length (bp)
help
|
2918318 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
140215 |
Topolino Chloroplast Genome
- Perianth: White
- Corona: yellow
- Division 1 (Trumpet daffodils)
- Hybridiser: J. Gerritsen & Son
- Registered: 1965
| Total Sequenced Bases (bp)
help
|
18,026,495 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
423.2 | N50 Read Length (bp)
help
|
466.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.62 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
2022 Daffodils
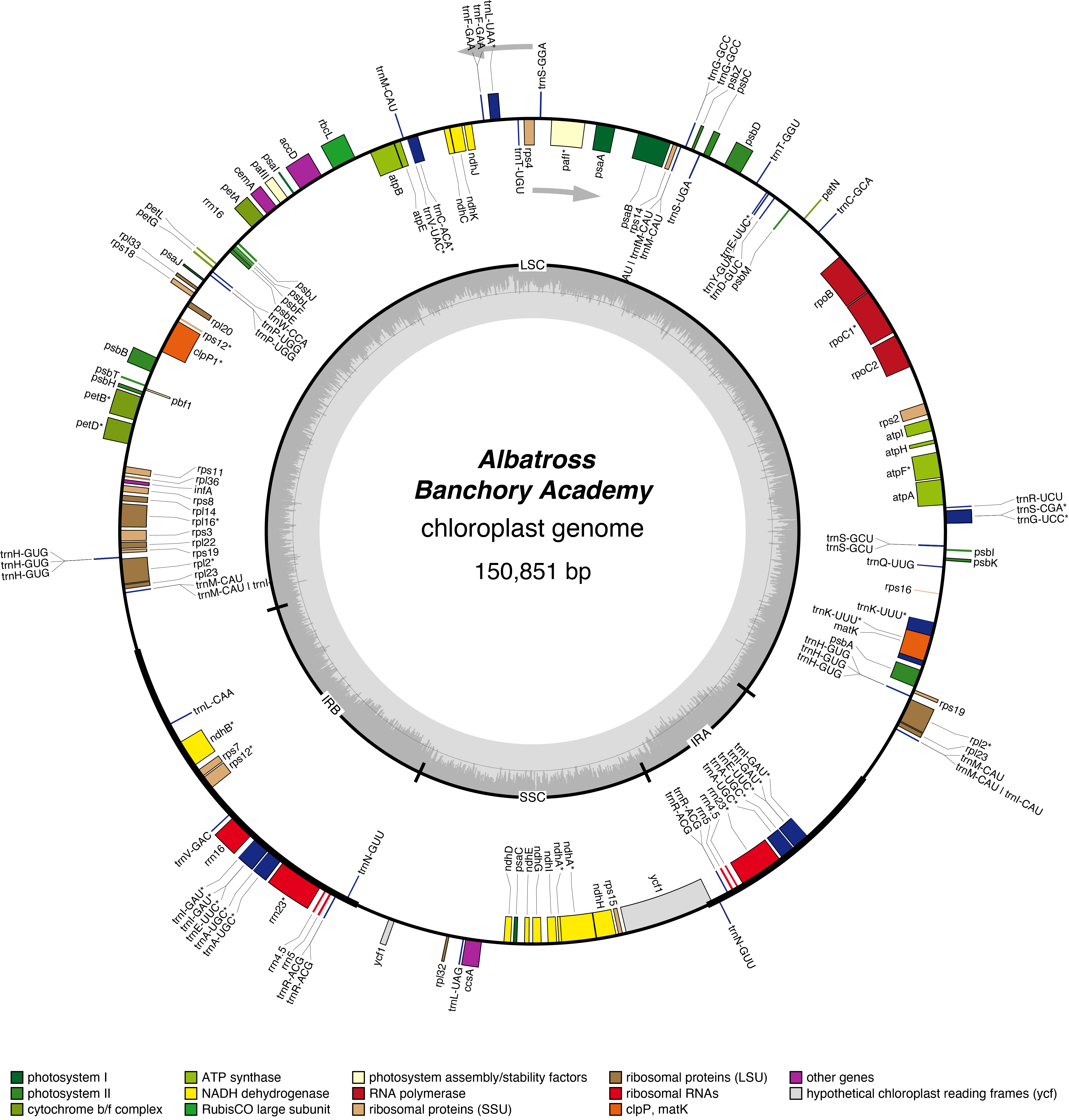
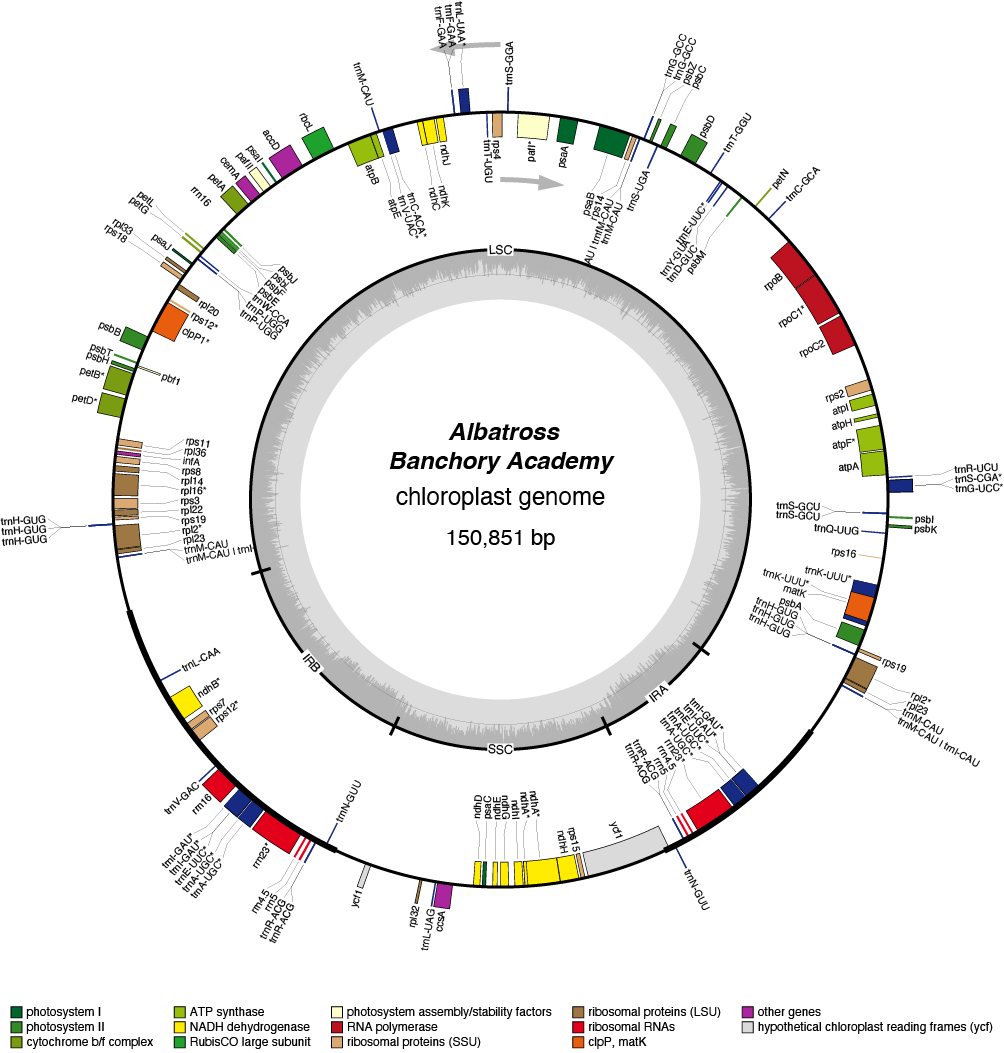
Albatross Chloroplast Genome
- Perianth: white
- Division 3 (Small cupped Daffodils)
- Hybridiser: Rev G. H. Englehart
- Registered: <1891

| Total Sequenced Bases (bp)
help
|
1,844,402,915 | Mean Quality
help
|
10.5 |
| Mean Read Length (bp)
help
|
2,408.6 | N50 Read Length (bp)
help
|
3,782.0 |
| Proportion of Chloroplast Bases (%)
help
|
2.8 | Chloroplast Coverage
help
|
327 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
150611 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
150759 |
Cheerio Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
148,406 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
1,613.1 | N50 Read Length (bp)
help
|
2,265.0 |
| Proportion of Chloroplast Bases (%)
help
|
11 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Coulmouny Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
941,653 | Mean Quality
help
|
10.1 |
| Mean Read Length (bp)
help
|
973.8 | N50 Read Length (bp)
help
|
1,556.0 |
| Proportion of Chloroplast Bases (%)
help
|
11 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Coverack beauty Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
689,139 | Mean Quality
help
|
10.1 |
| Mean Read Length (bp)
help
|
1,027.0 | N50 Read Length (bp)
help
|
1,512.0 |
| Proportion of Chloroplast Bases (%)
help
|
16 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
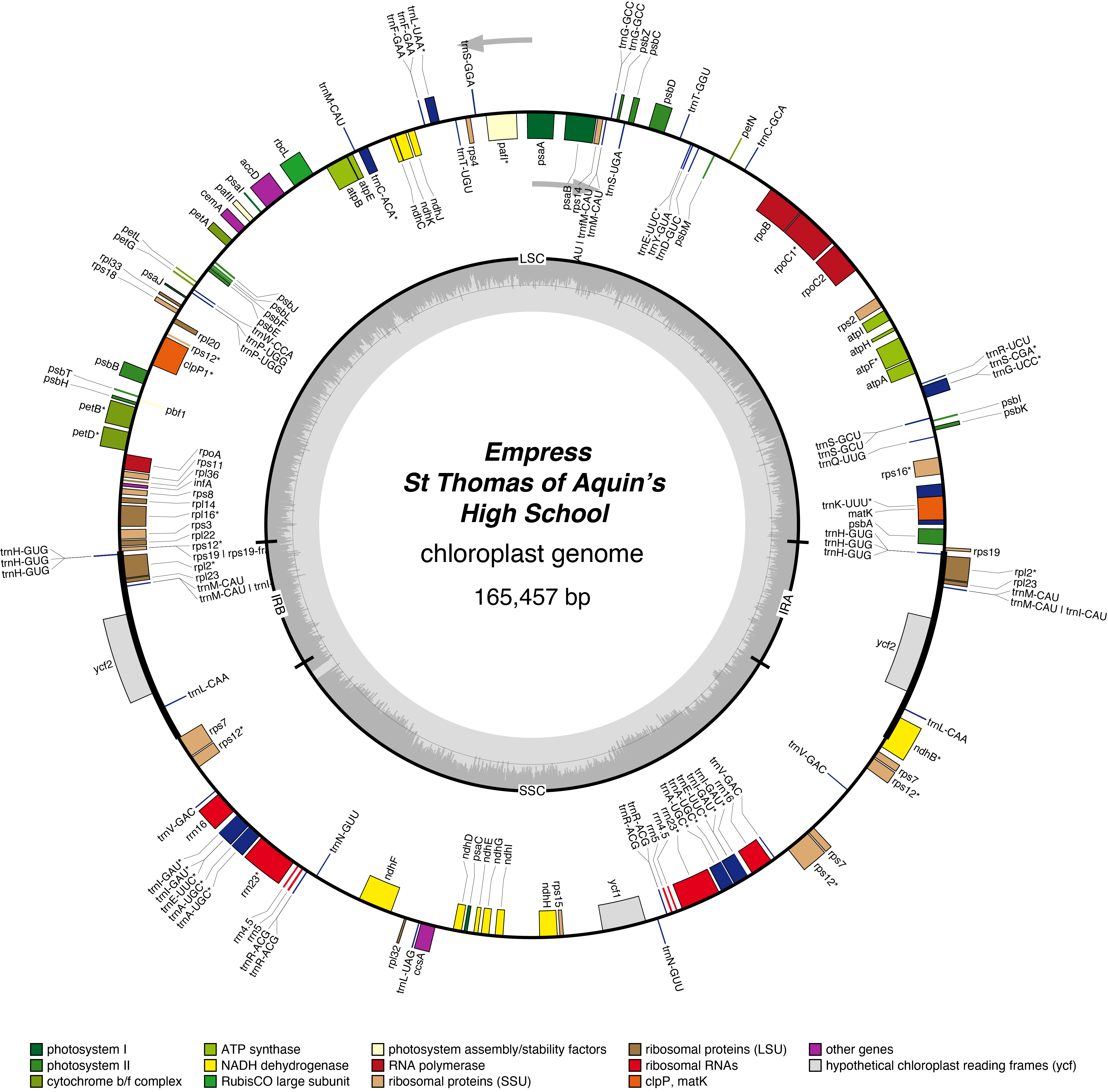
Empress Chloroplast Genome
- Corona: yellow
- Perianth: white
- Division: 1 (Trumpet Daffodils)
- Height: Standard (32-68cm)
- Hybridiser: William Backhouse
- Registered: <1869

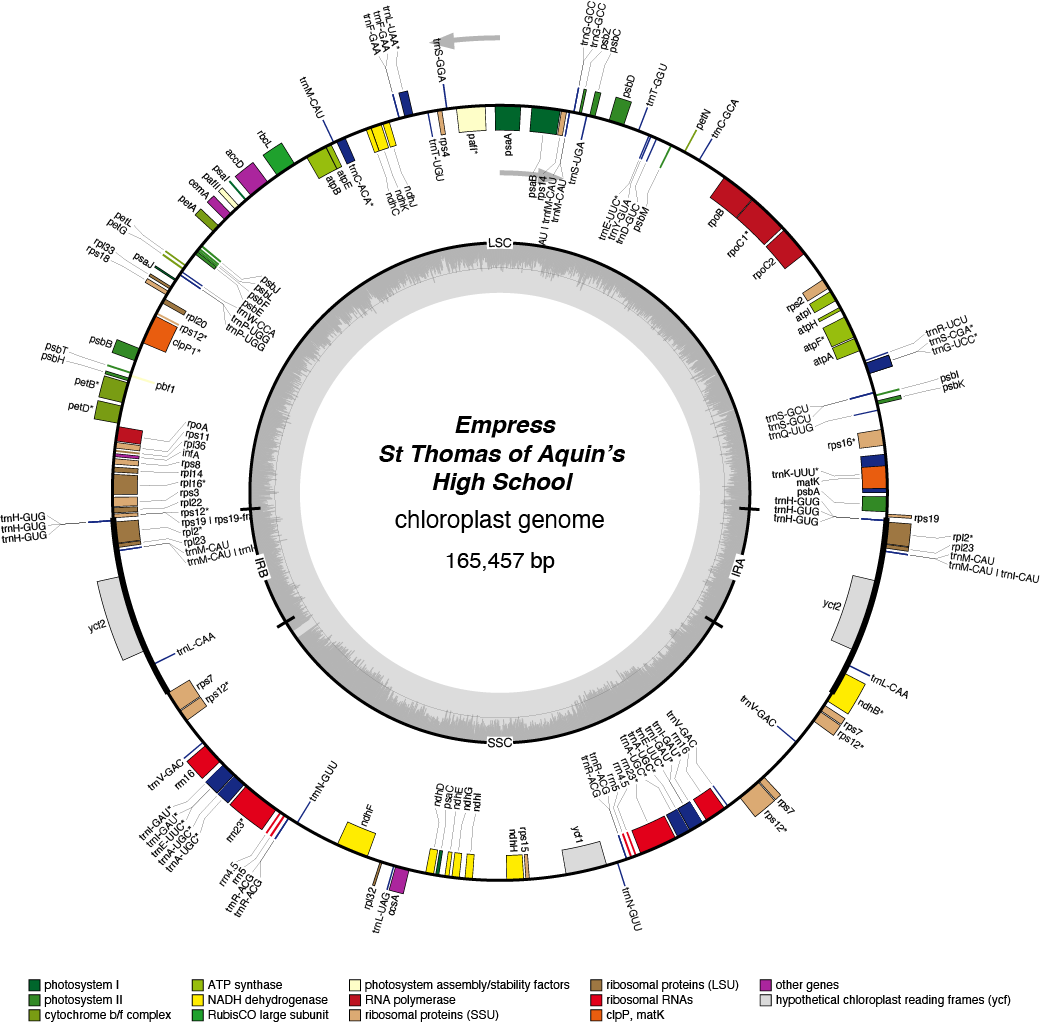
| Total Sequenced Bases (bp)
help
|
154,483,778 | Mean Quality
help
|
11.3 |
| Mean Read Length (bp)
help
|
1,906.5 | N50 Read Length (bp)
help
|
3,539.0 |
| Proportion of Chloroplast Bases (%)
help
|
17 | Chloroplast Coverage
help
|
165 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
164568 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
165506 |
Forest Fire Chloroplast Genome
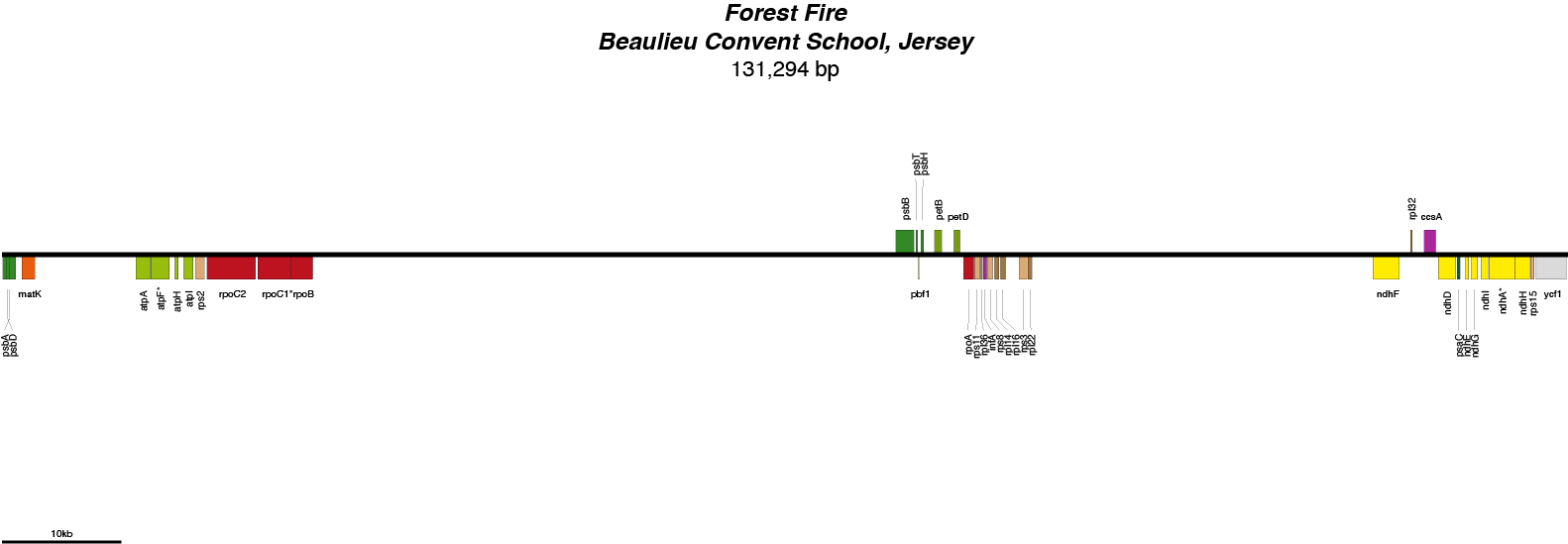
| Total Sequenced Bases (bp)
help
|
16,703,346 | Mean Quality
help
|
10.0 |
| Mean Read Length (bp)
help
|
2,058.6 | N50 Read Length (bp)
help
|
4,184.0 |
| Proportion of Chloroplast Bases (%)
help
|
3.3 | Chloroplast Coverage
help
|
3 |
| Assembled Contigs
help
|
4 | Assembled Contig Length (bp)
help
|
45827 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
131294 |
Furness Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
660,329 | Mean Quality
help
|
11.0 |
| Mean Read Length (bp)
help
|
2,934.8 | N50 Read Length (bp)
help
|
6,953.0 |
| Proportion of Chloroplast Bases (%)
help
|
5 | Chloroplast Coverage
help
|
0 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
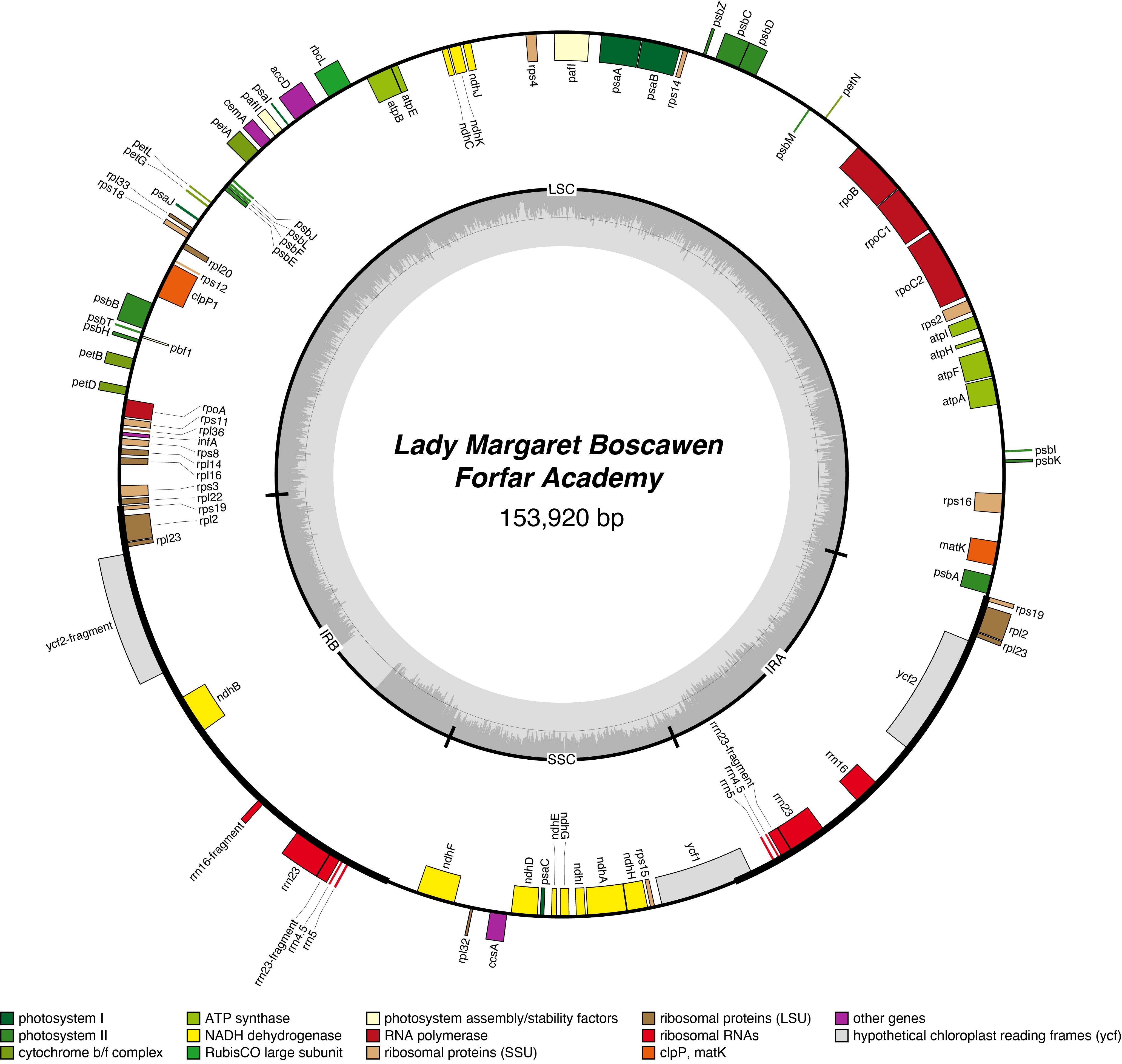
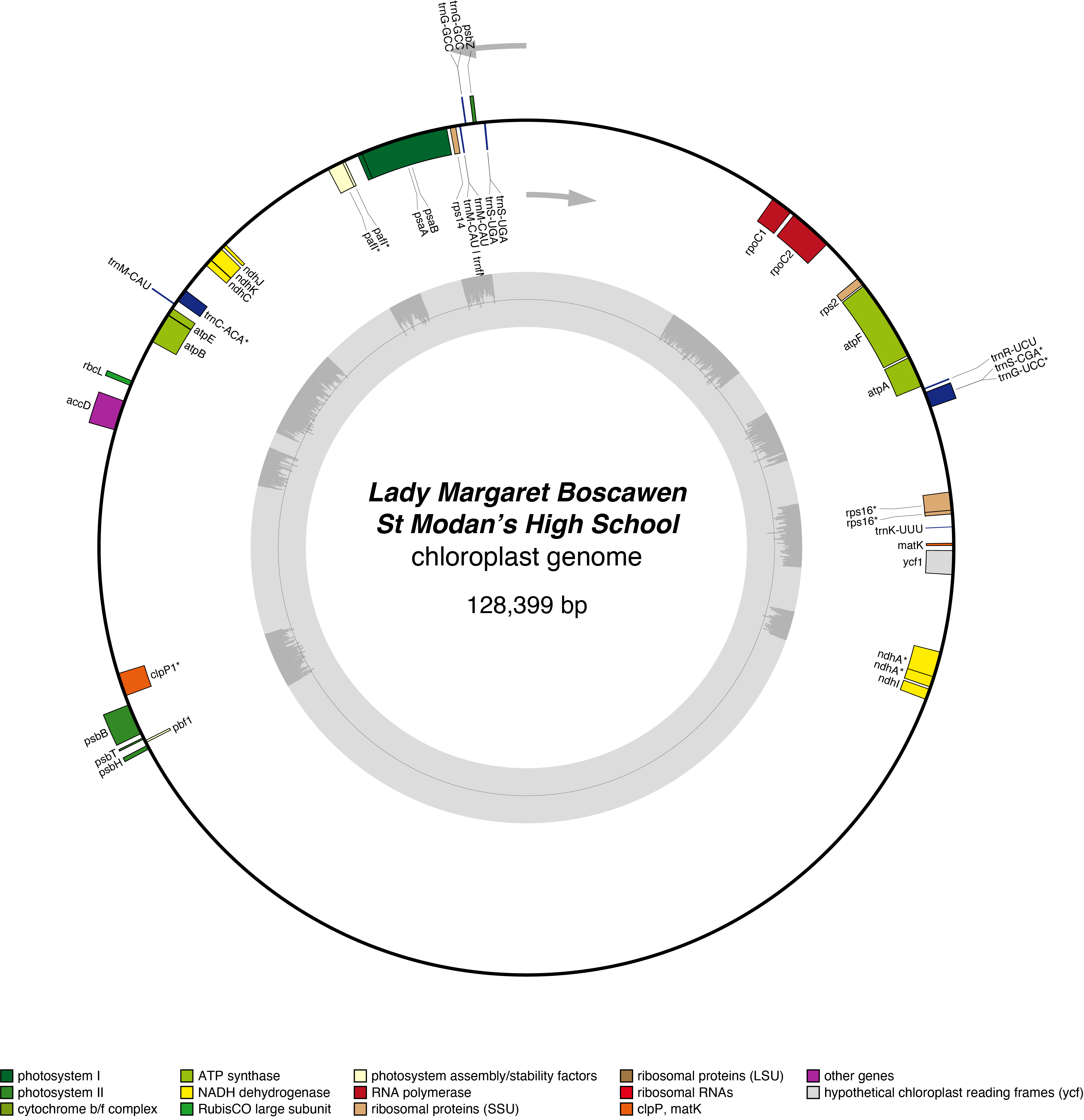
Lady Margaret Boscawen Chloroplast Genome
- Corona: yellow
- Perianth: white
- Division: 2 (Large-Cupped Daffodils)
- Hybridiser: G.H. Engleheart
- Registered: <1898

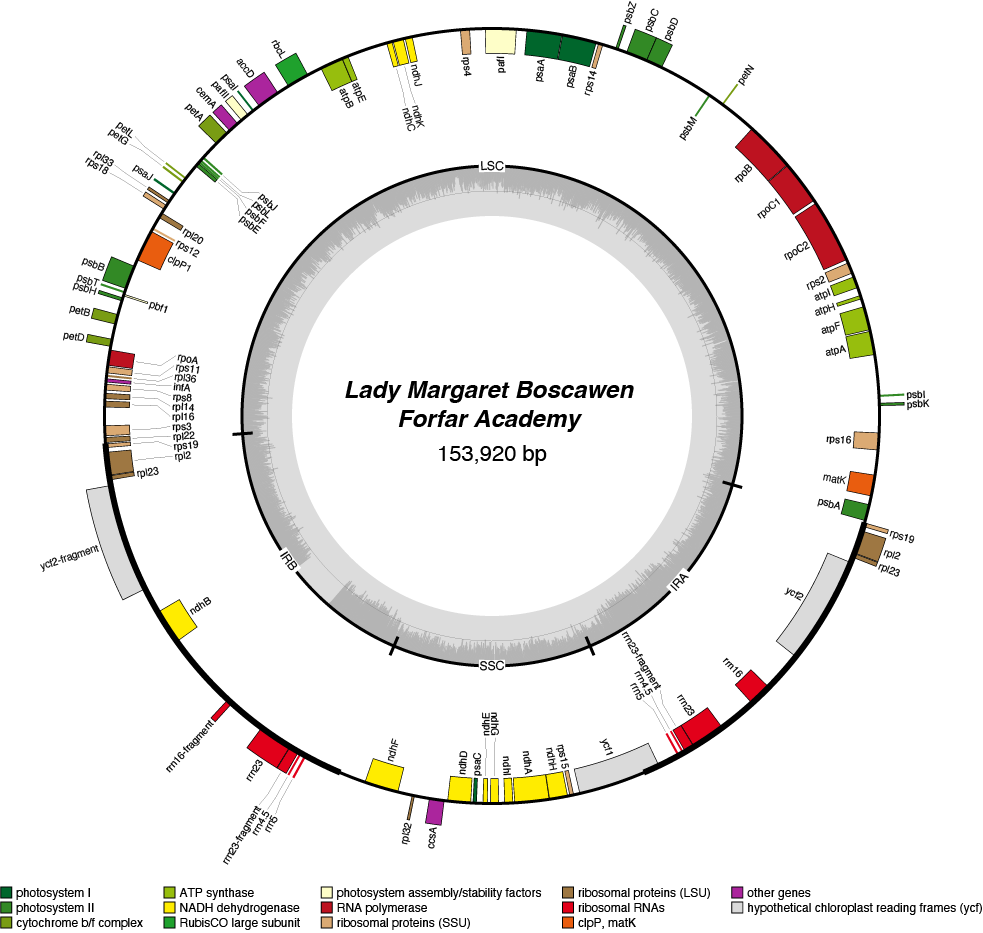
| Total Sequenced Bases (bp)
help
|
1,293,024,147 | Mean Quality
help
|
12.1 |
| Mean Read Length (bp)
help
|
1,072.8 | N50 Read Length (bp)
help
|
1,558.0 |
| Proportion of Chloroplast Bases (%)
help
|
37 | Chloroplast Coverage
help
|
2,982 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
149075 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
153893 |
Lady Margaret Boscawen Chloroplast Genome
- Corona: yellow
- Perianth: white
- Division: 2 (Large-Cupped Daffodils)
- Hybridiser: G.H. Engleheart
- Registered: <1898

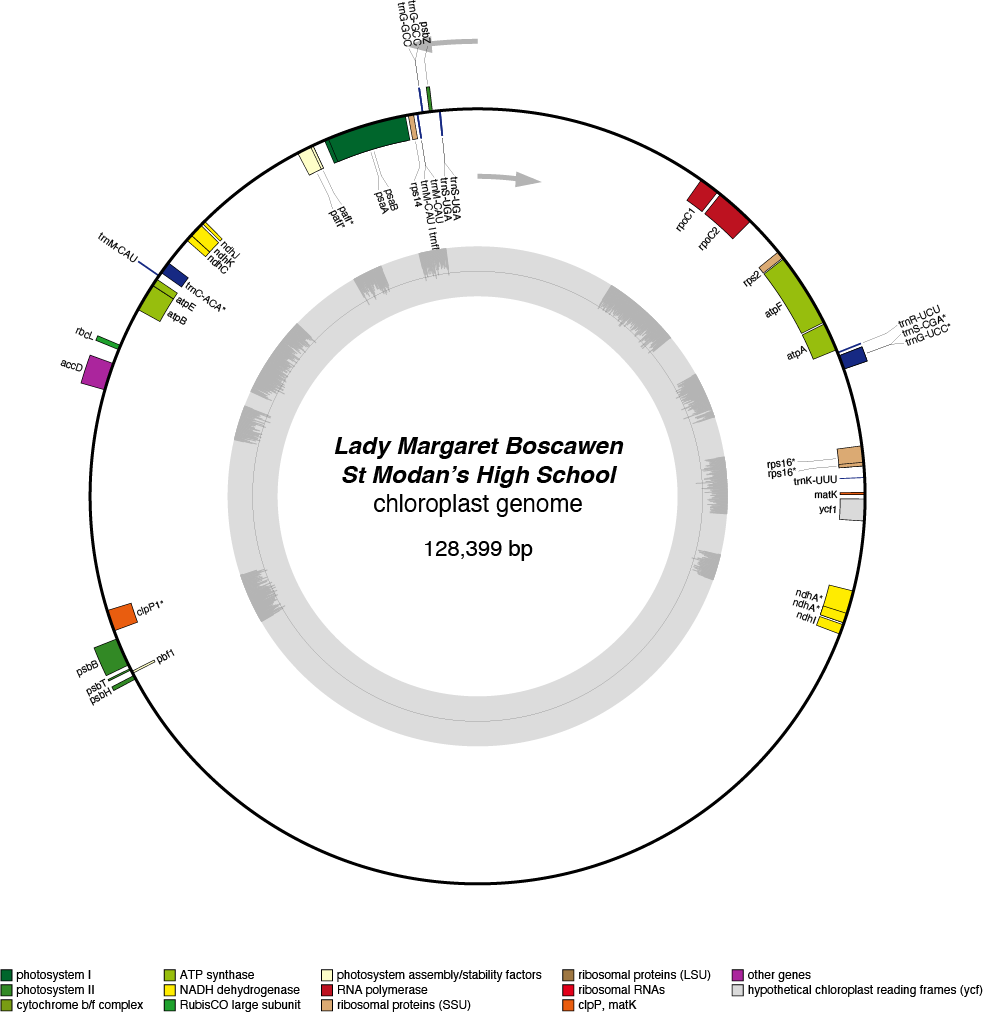
| Total Sequenced Bases (bp)
help
|
43,051,165 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
1,531.3 | N50 Read Length (bp)
help
|
2,428.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.5 | Chloroplast Coverage
help
|
3 |
| Assembled Contigs
help
|
10 | Assembled Contig Length (bp)
help
|
35625 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
128305 |
Loch Fyne Chloroplast Genome
- Corona: yellow
- Perianth: white
- Division: 2 (Large-Cupped Daffodils)
- Hybridiser: The Brodie of Brodie
- Registered: <1911

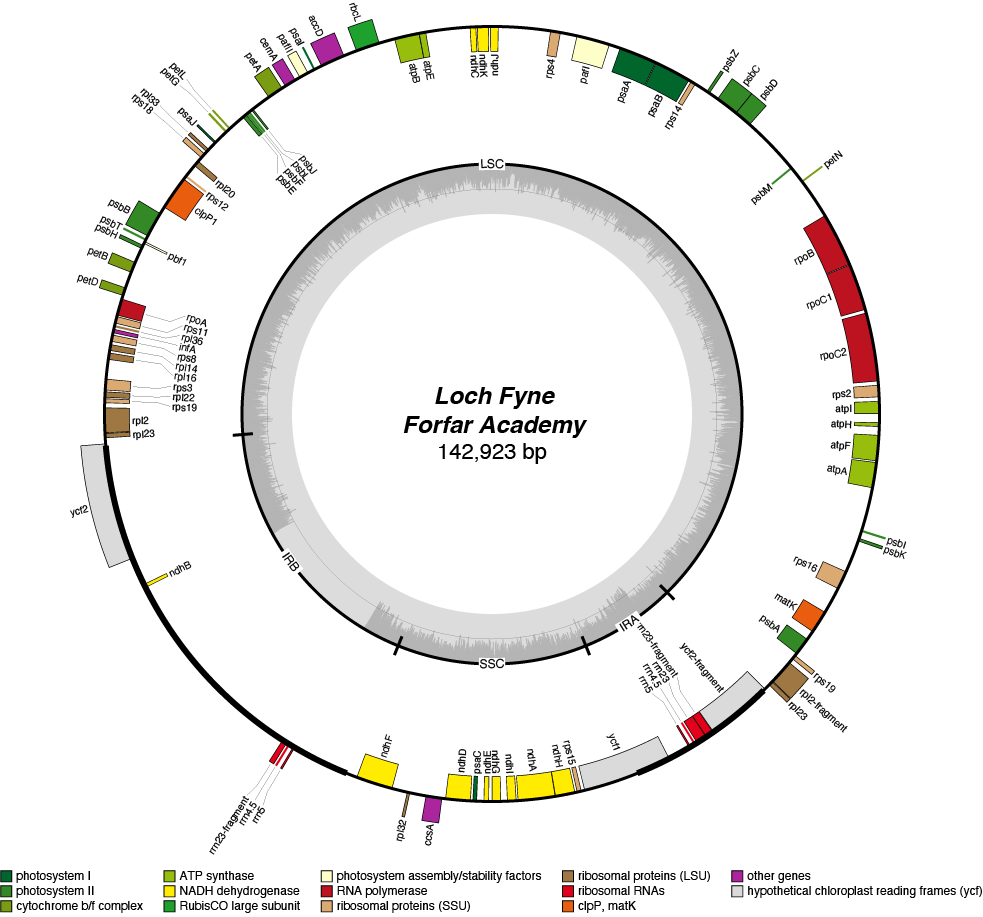
| Total Sequenced Bases (bp)
help
|
276,232,033 | Mean Quality
help
|
11.9 |
| Mean Read Length (bp)
help
|
751.6 | N50 Read Length (bp)
help
|
1,024.0 |
| Proportion of Chloroplast Bases (%)
help
|
55 | Chloroplast Coverage
help
|
954 |
| Assembled Contigs
help
|
10 | Assembled Contig Length (bp)
help
|
184993 |
| Assembled Scaffolds
help
|
2 | Assembled Scaffold Length (bp)
help
|
208919 |
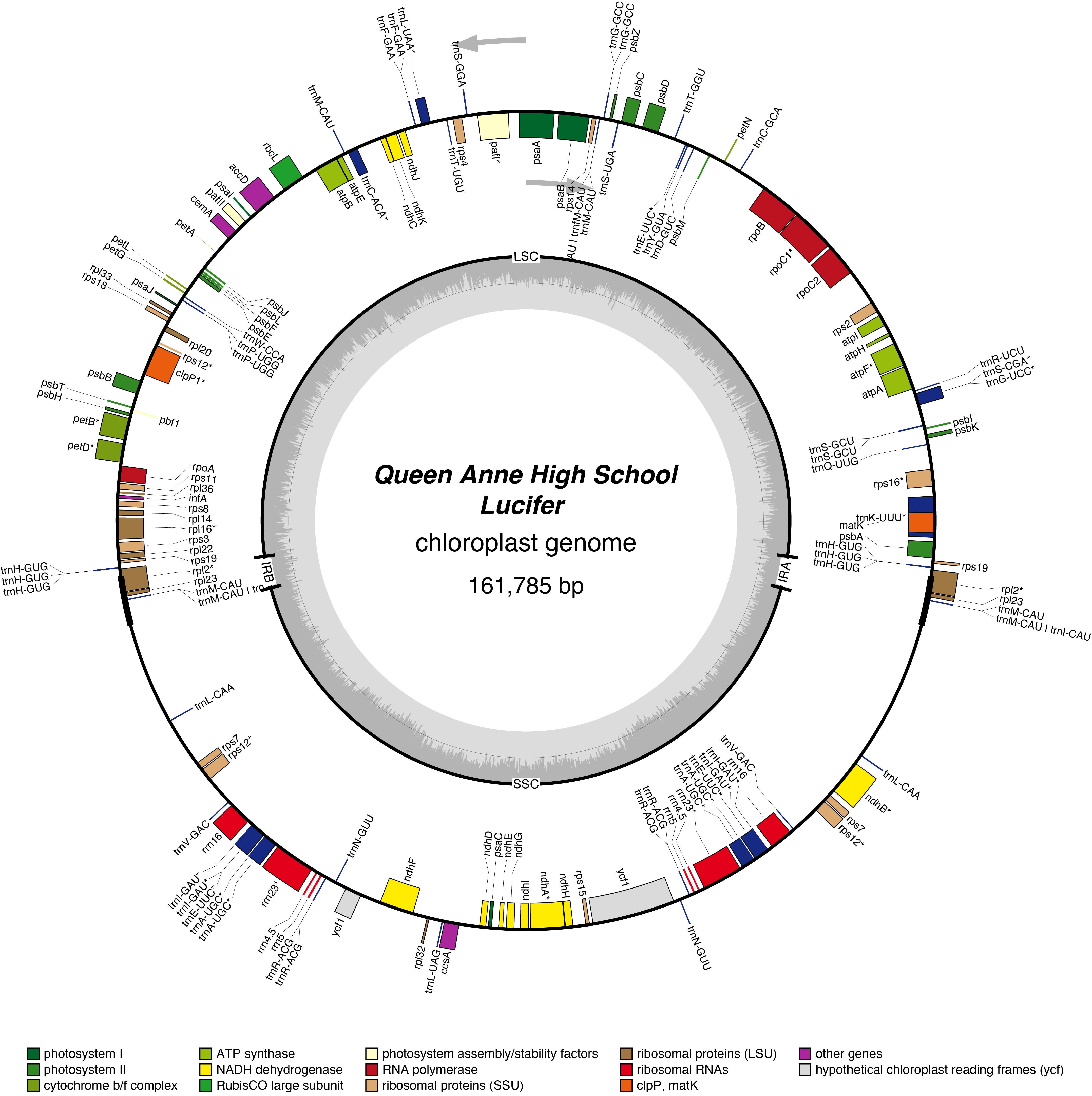
Lucifer Chloroplast Genome
- Corona: orange
- Perianth: white
- Division: 3 (Small-Cupped Daffodils)
- Hybridiser: G.H. Engleheart
- Registered: <1897

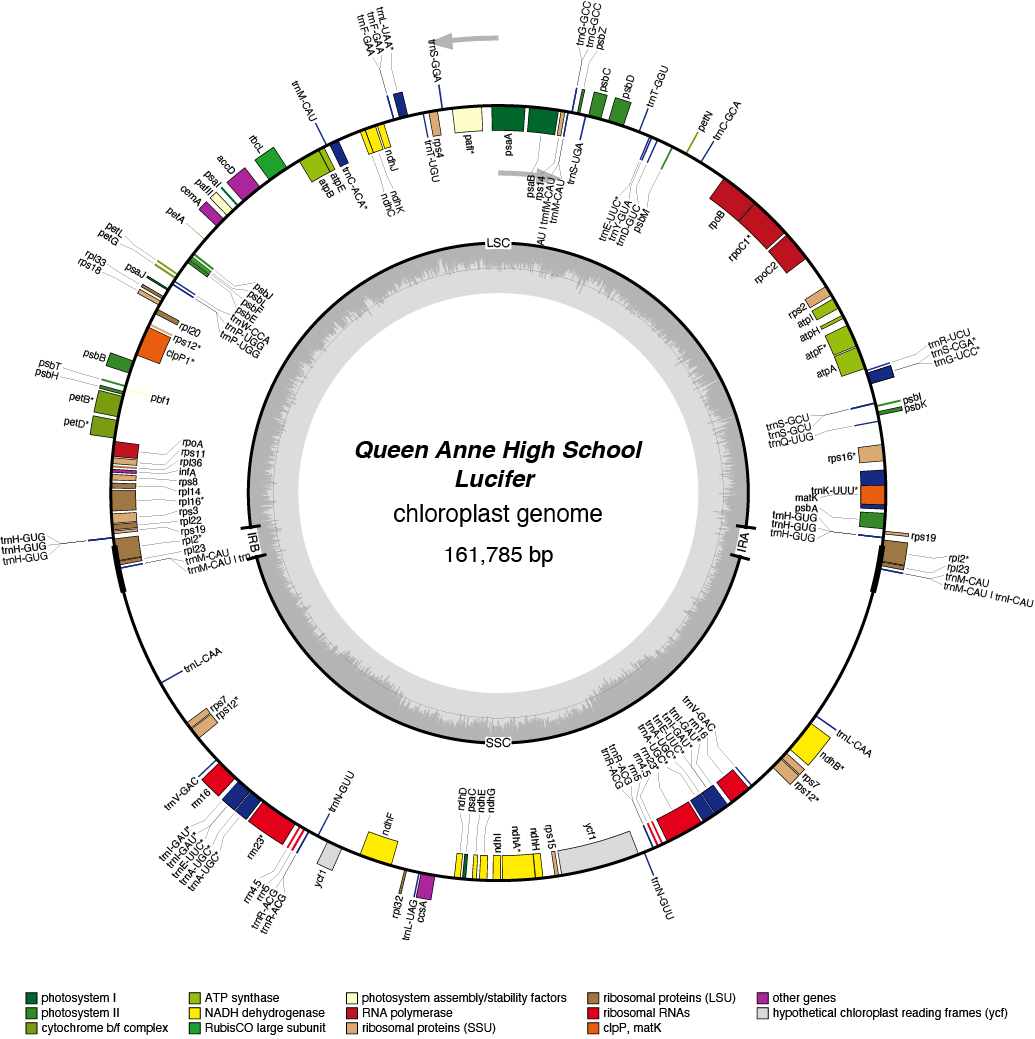
| Total Sequenced Bases (bp)
help
|
3,793,871,703 | Mean Quality
help
|
10.7 |
| Mean Read Length (bp)
help
|
2,344.6 | N50 Read Length (bp)
help
|
4,325.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.9 | Chloroplast Coverage
help
|
451 |
| Assembled Contigs
help
|
1 | Assembled Contig Length (bp)
help
|
161992 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
161992 |
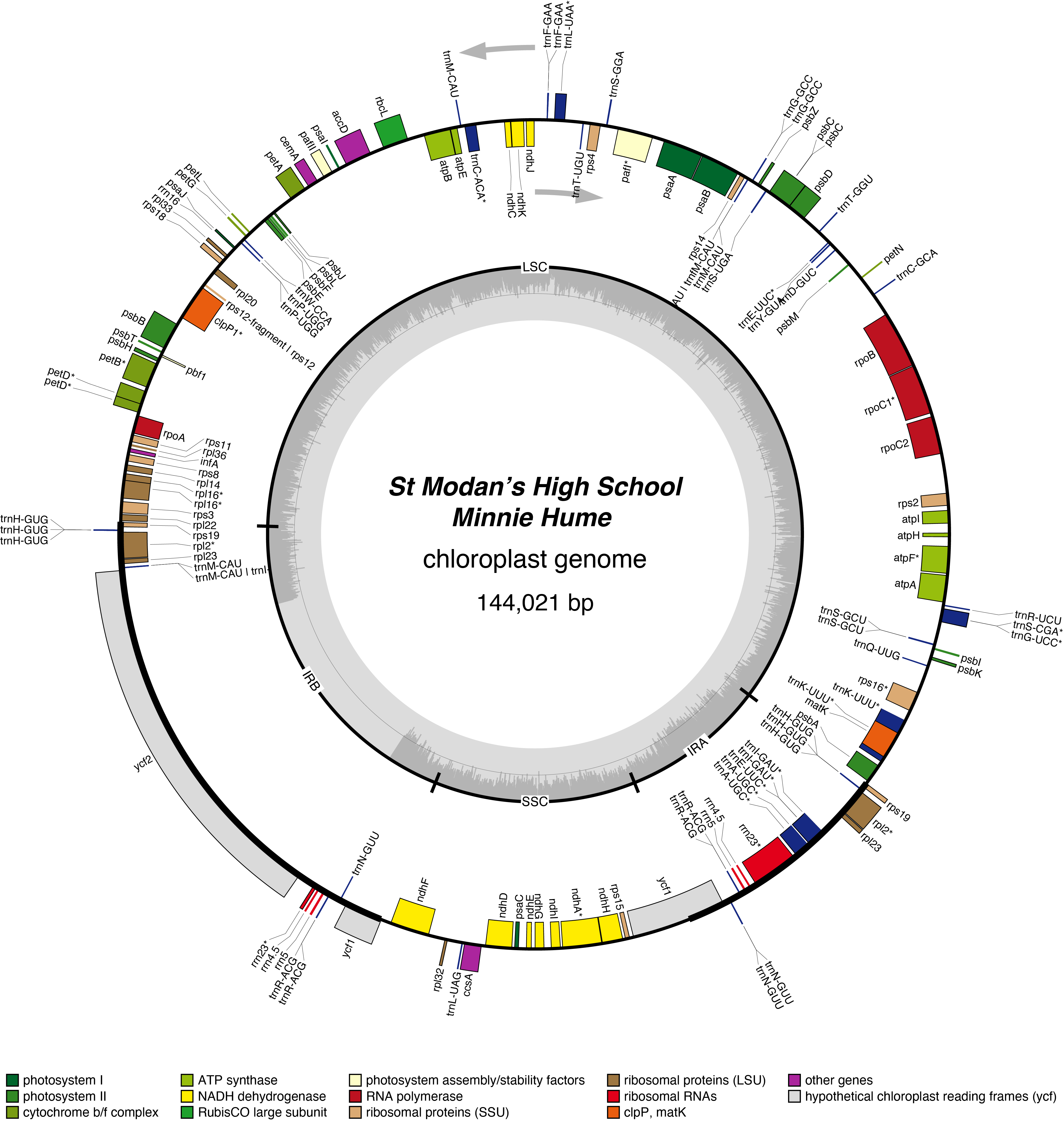
Minnie Hume Chloroplast Genome
- Corona: white
- Perianth: white
- Division: 3 (Small-Cupped Daffodils)
- Hybridiser: William Backhouse
- Registered: <1889

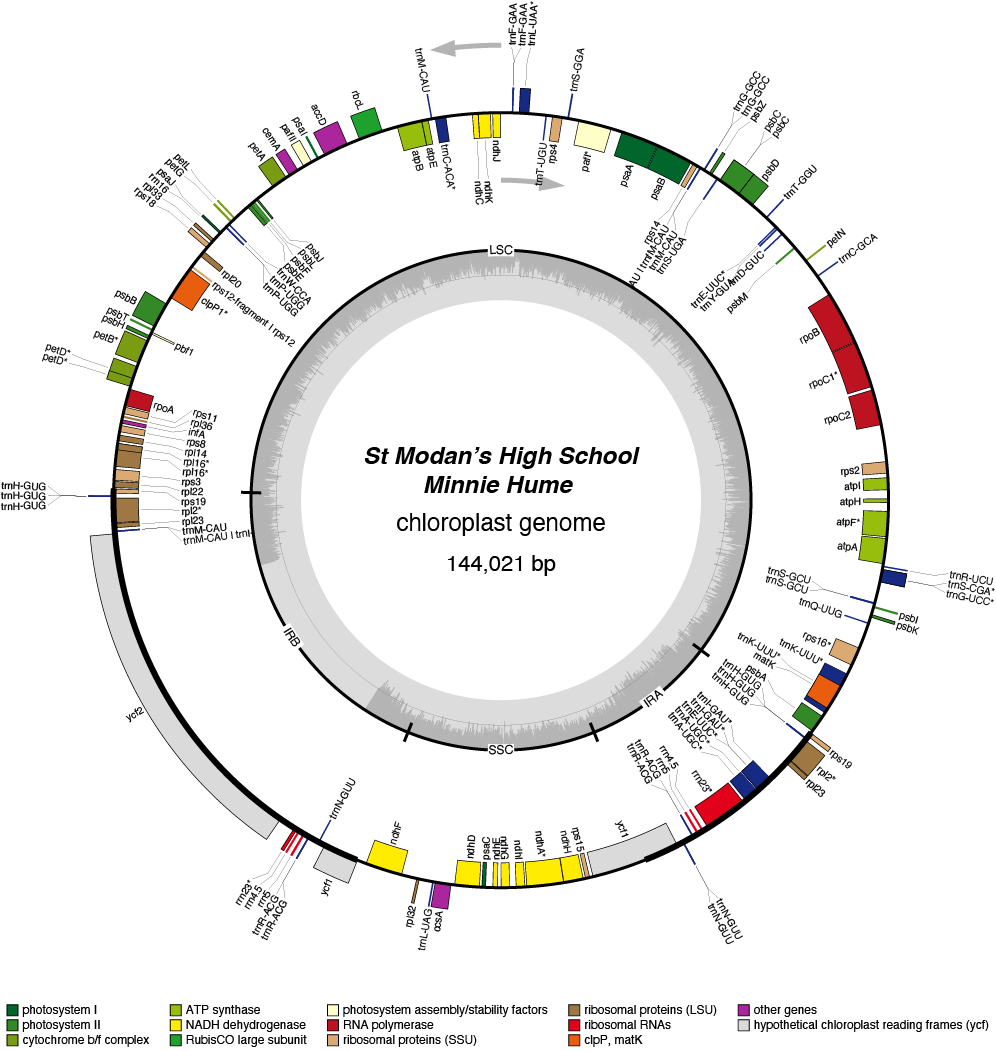
| Total Sequenced Bases (bp)
help
|
637,388,752 | Mean Quality
help
|
10.3 |
| Mean Read Length (bp)
help
|
1,562.3 | N50 Read Length (bp)
help
|
2,449.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.7 | Chloroplast Coverage
help
|
65 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
127041 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
143789 |
Morven Chloroplast Genome
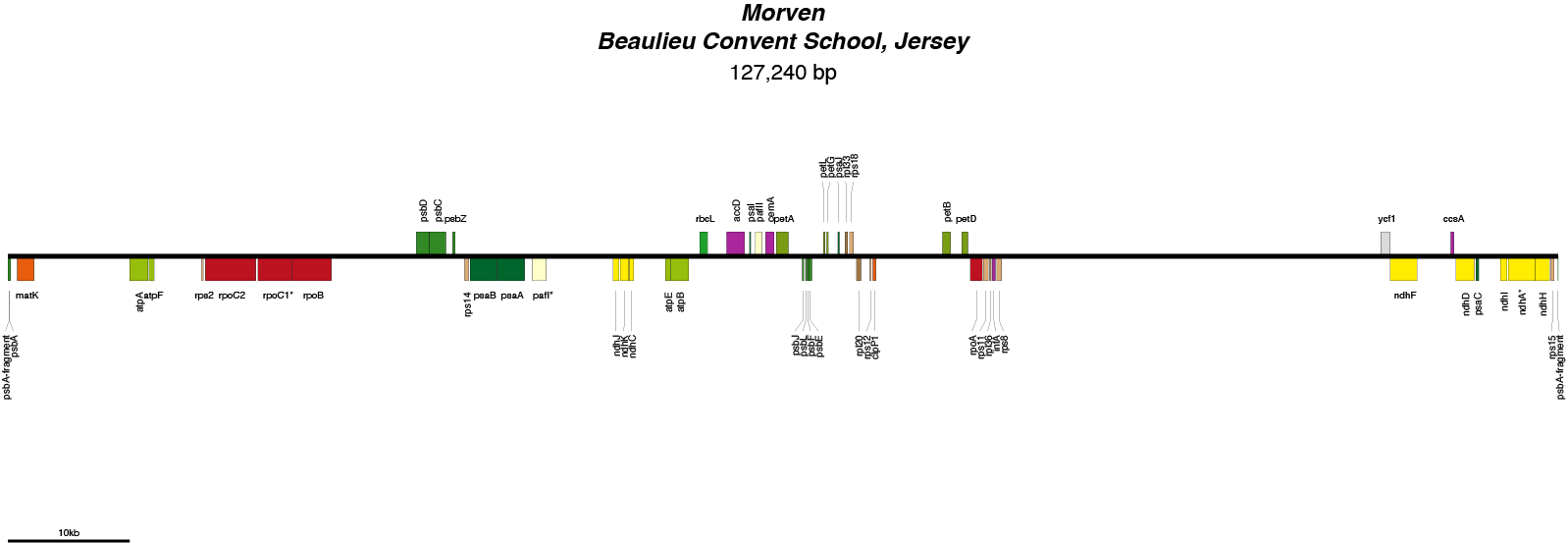
| Total Sequenced Bases (bp)
help
|
11,975,706 | Mean Quality
help
|
10.4 |
| Mean Read Length (bp)
help
|
1,217.8 | N50 Read Length (bp)
help
|
2,056.0 |
| Proportion of Chloroplast Bases (%)
help
|
9 | Chloroplast Coverage
help
|
6 |
| Assembled Contigs
help
|
12 | Assembled Contig Length (bp)
help
|
70605 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
127240 |
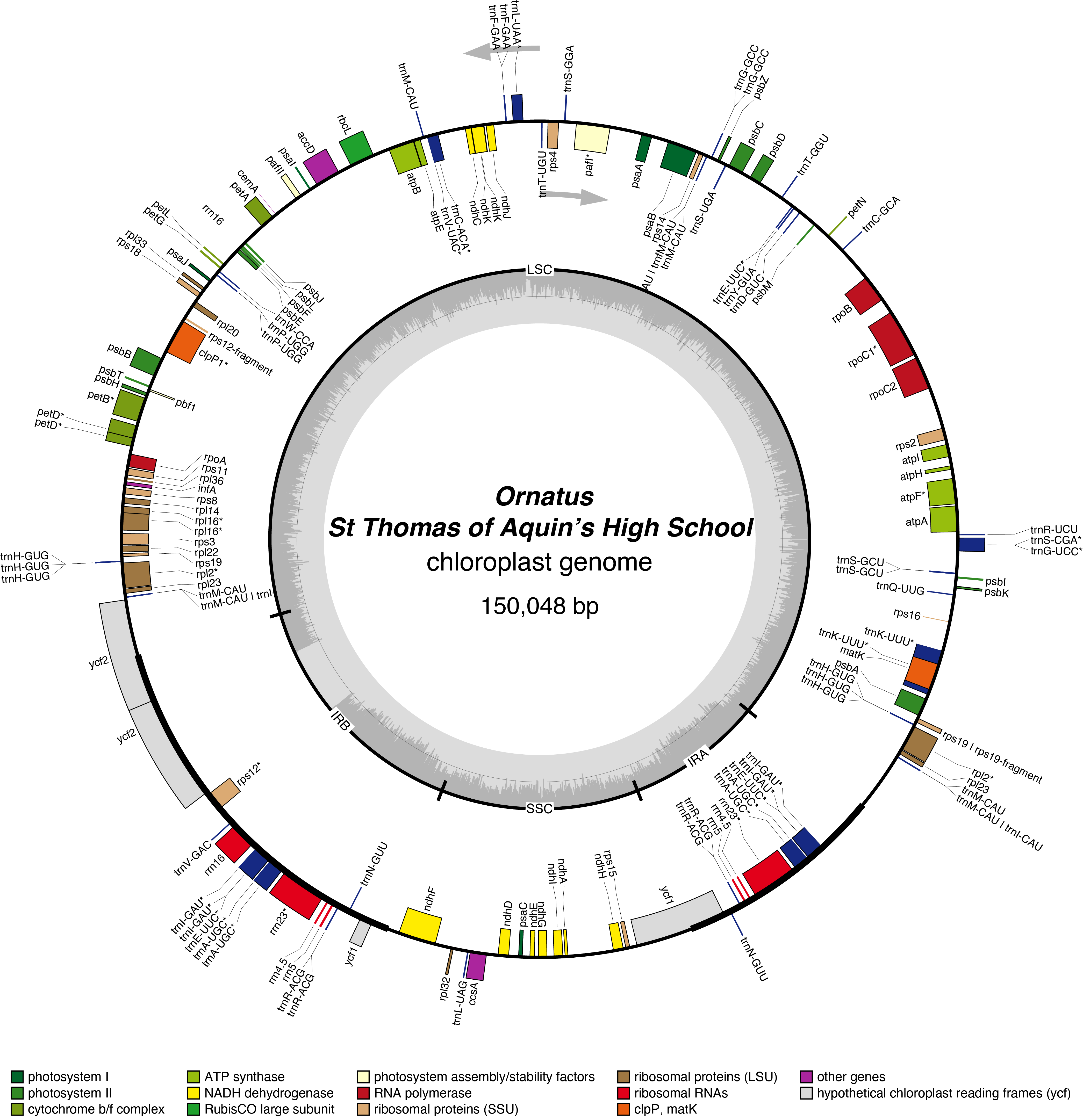
Ornatus Chloroplast Genome
- Perianth: white
- Division 9 (Poeticus Daffodils)
- Selected: James Walker
- Registered: <1870

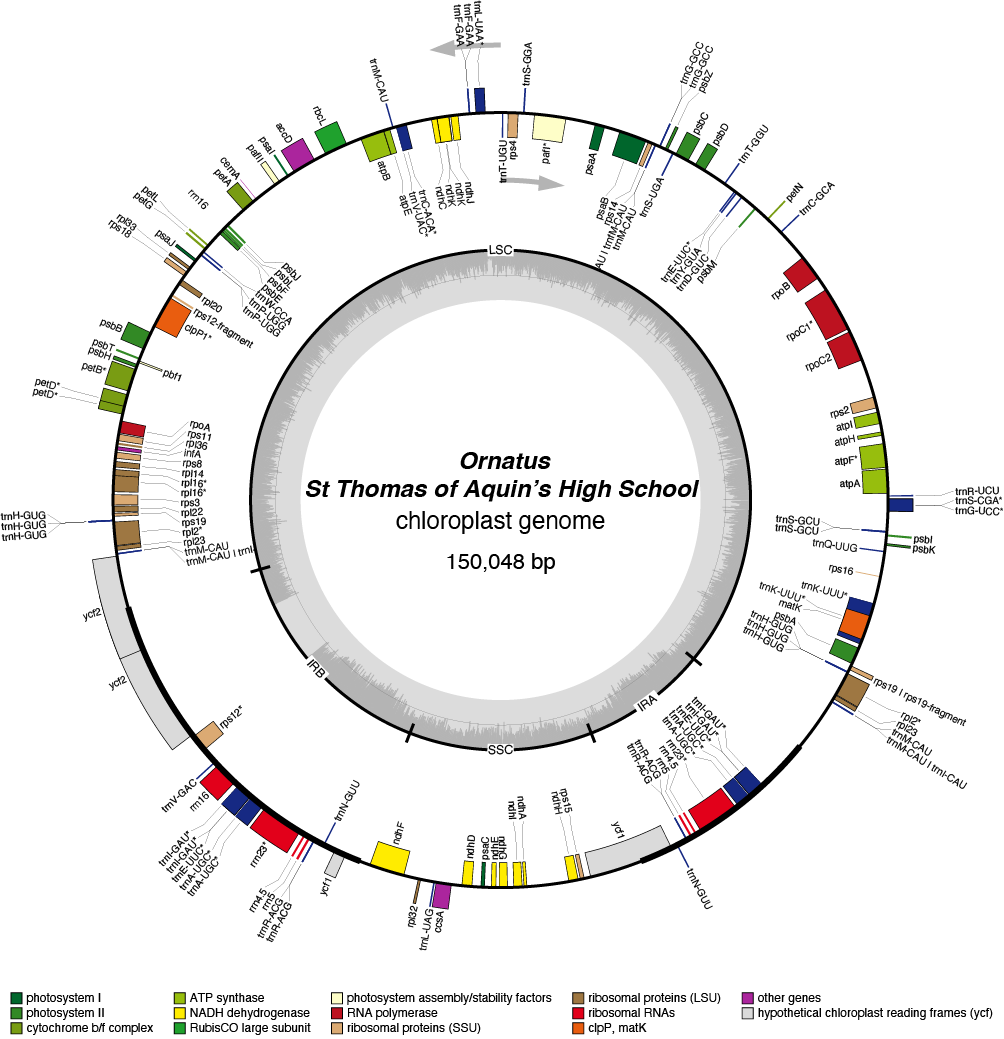
| Total Sequenced Bases (bp)
help
|
172,644,669 | Mean Quality
help
|
10.3 |
| Mean Read Length (bp)
help
|
2,731.9 | N50 Read Length (bp)
help
|
4,175.0 |
| Proportion of Chloroplast Bases (%)
help
|
9.9 | Chloroplast Coverage
help
|
106 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
143892 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
149951 |
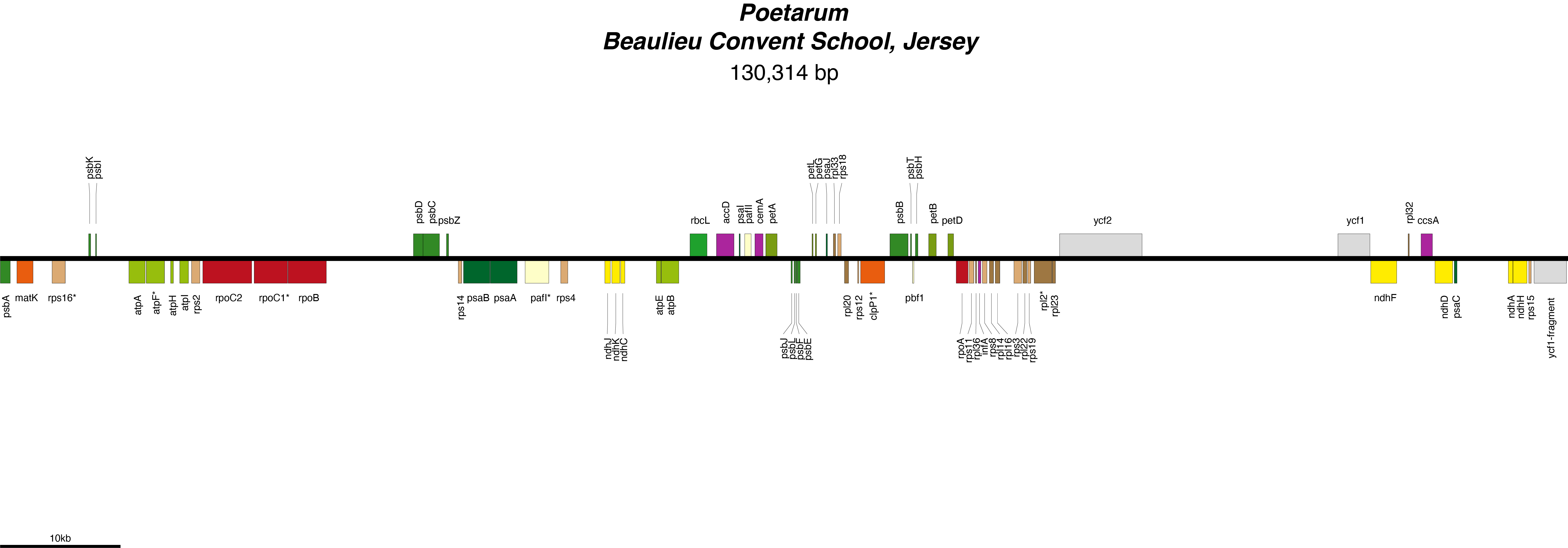
Poetarum Chloroplast Genome
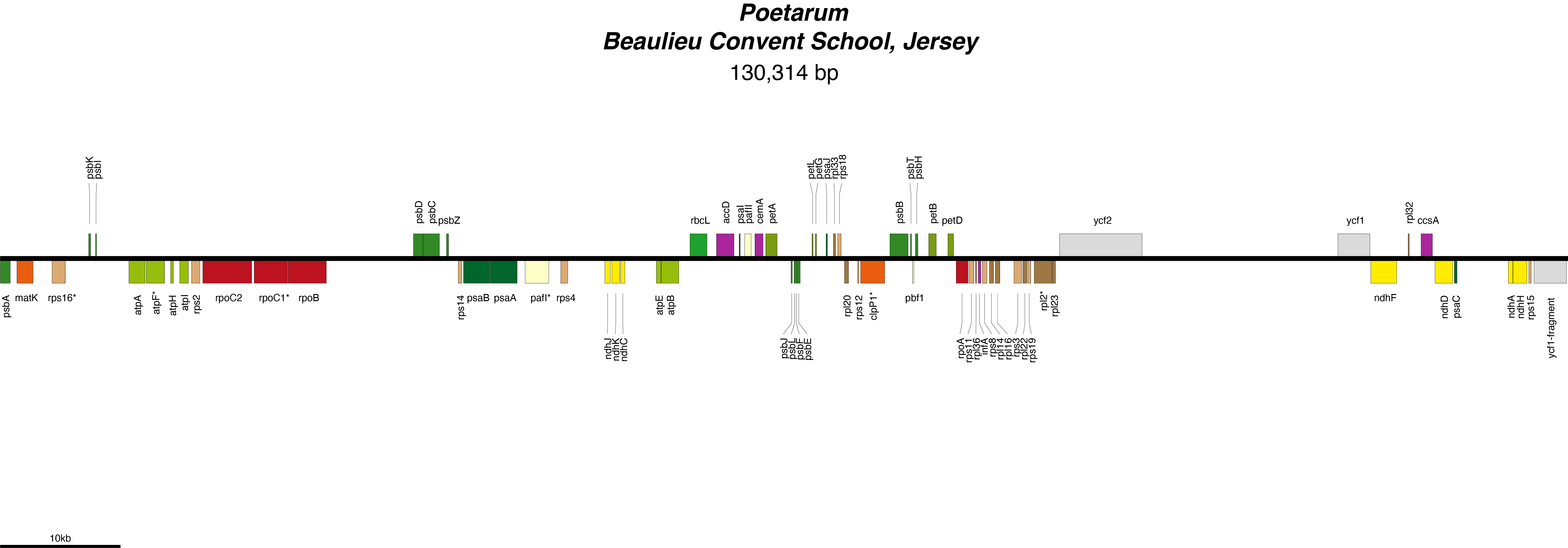
| Total Sequenced Bases (bp)
help
|
13,958,953 | Mean Quality
help
|
10.3 |
| Mean Read Length (bp)
help
|
2,510.2 | N50 Read Length (bp)
help
|
5,812.0 |
| Proportion of Chloroplast Bases (%)
help
|
6.9 | Chloroplast Coverage
help
|
6 |
| Assembled Contigs
help
|
6 | Assembled Contig Length (bp)
help
|
105128 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
130314 |
Poetarum Chloroplast Genome
- Perianth: white
- Division 13 - Species, Wild Variants

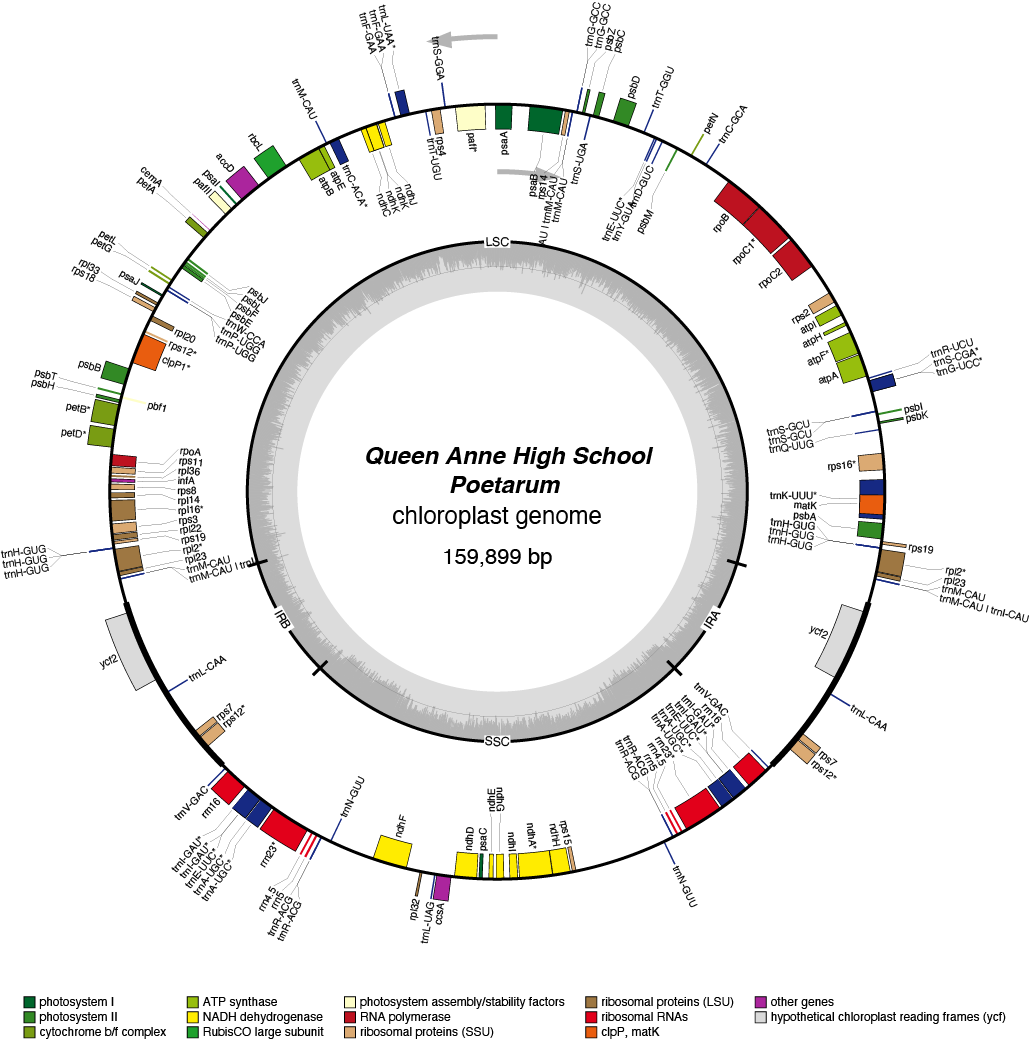
| Total Sequenced Bases (bp)
help
|
4,063,677,562 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
3,396.6 | N50 Read Length (bp)
help
|
5,846.0 |
| Proportion of Chloroplast Bases (%)
help
|
1.4 | Chloroplast Coverage
help
|
353 |
| Assembled Contigs
help
|
2 | Assembled Contig Length (bp)
help
|
159765 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
159865 |
Princeps Chloroplast Genome
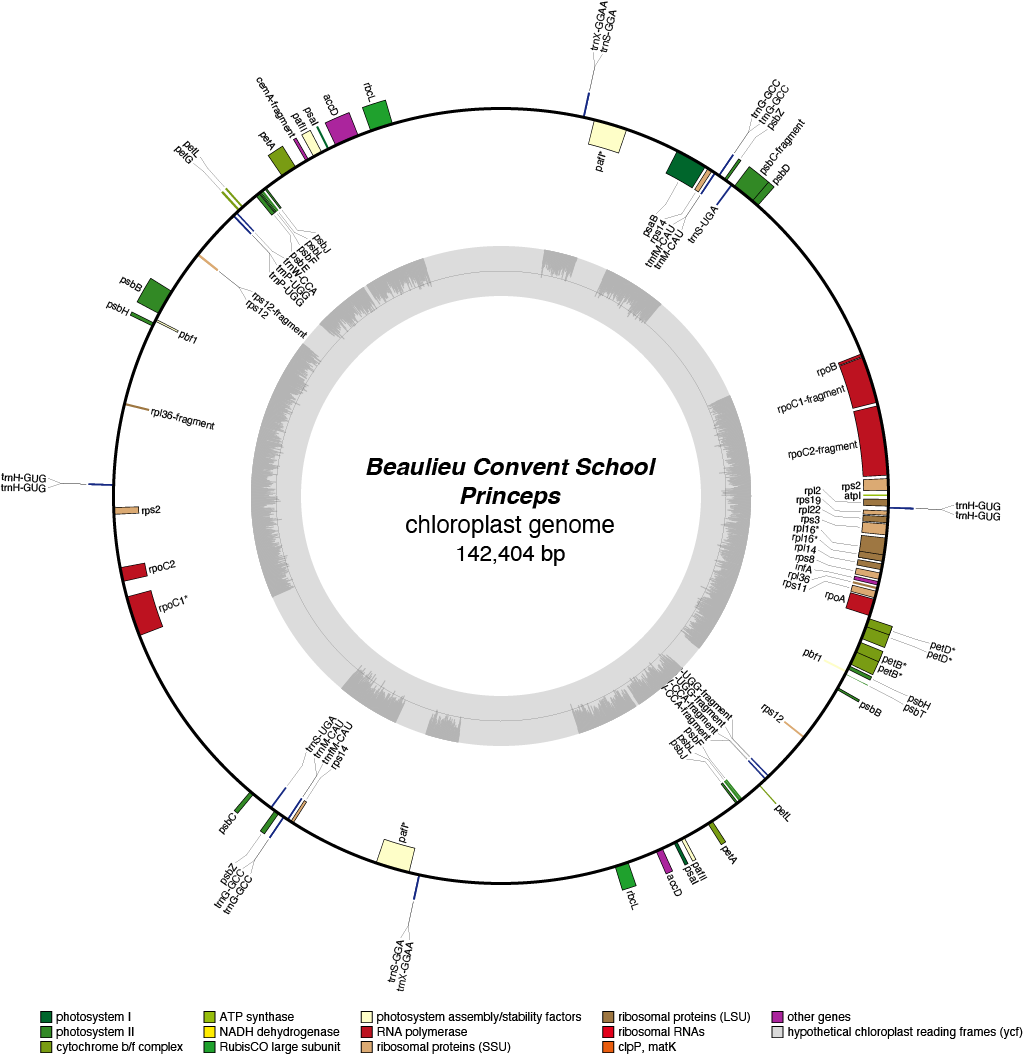
| Total Sequenced Bases (bp)
help
|
4,591,656 | Mean Quality
help
|
10.1 |
| Mean Read Length (bp)
help
|
912.5 | N50 Read Length (bp)
help
|
1,442.0 |
| Proportion of Chloroplast Bases (%)
help
|
12 | Chloroplast Coverage
help
|
3 |
| Assembled Contigs
help
|
6 | Assembled Contig Length (bp)
help
|
43778 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
142227 |
Princeps Chloroplast Genome
- Perianth: white
- Division 1 (Trumpet Daffodils)
- Hybridiser: Unknown
- Registered: <1830

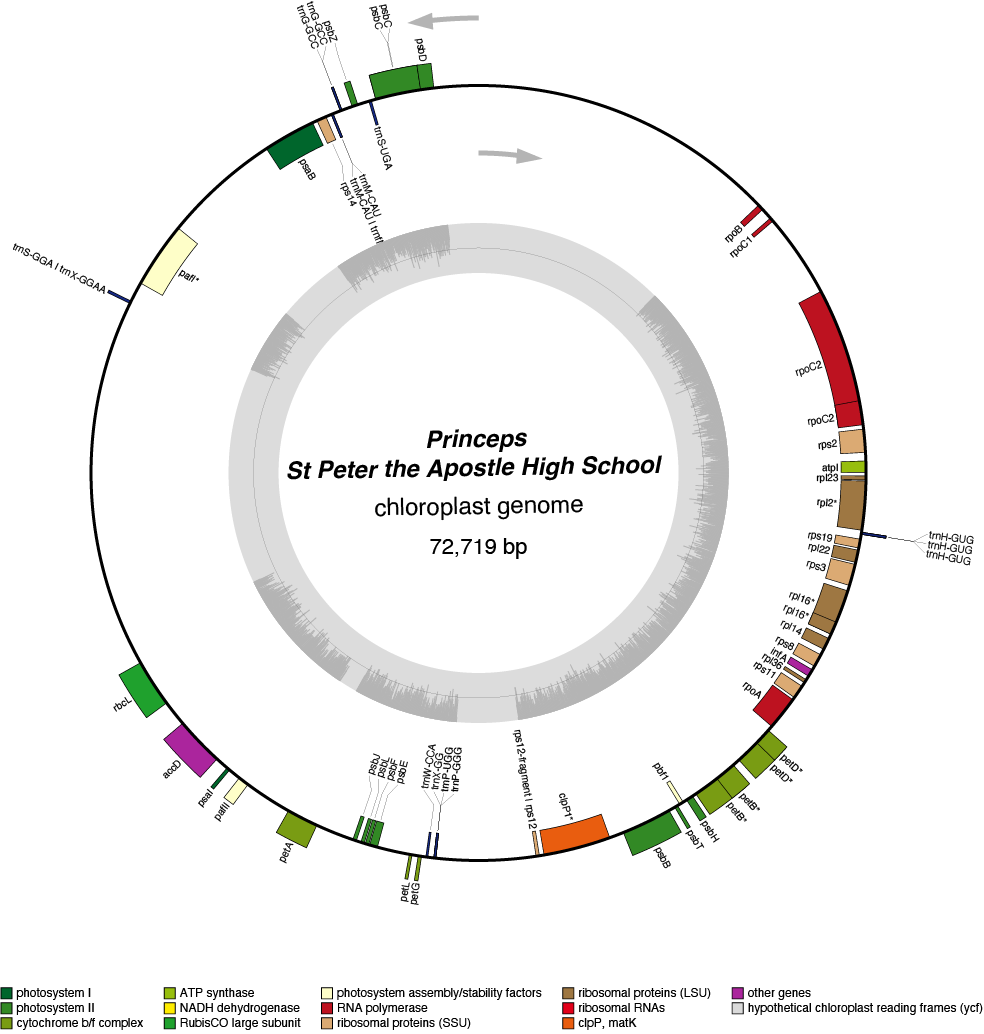
| Total Sequenced Bases (bp)
help
|
558,705,420 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
740.5 | N50 Read Length (bp)
help
|
1,020.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.13 | Chloroplast Coverage
help
|
4 |
| Assembled Contigs
help
|
6 | Assembled Contig Length (bp)
help
|
44844 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
72568 |
Princeps Chloroplast Genome
- Perianth: white
- Division 1 (Trumpet Daffodils)
- Hybridiser: Unknown
- Registered: <1830

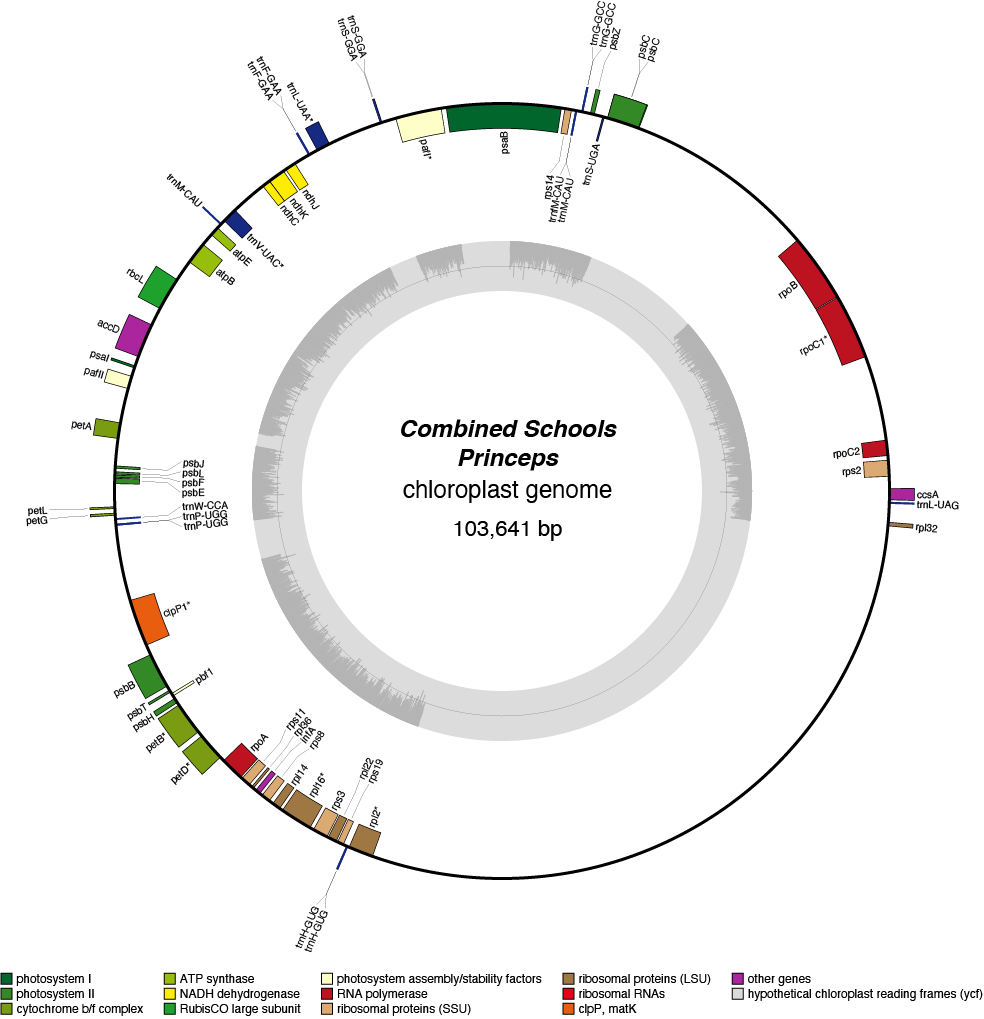
| Total Sequenced Bases (bp)
help
|
615,978,115 | Mean Quality
help
|
10.2 |
| Mean Read Length (bp)
help
|
744.4 | N50 Read Length (bp)
help
|
1,014.0 |
| Proportion of Chloroplast Bases (%)
help
|
0.18 | Chloroplast Coverage
help
|
6 |
| Assembled Contigs
help
|
8 | Assembled Contig Length (bp)
help
|
57237 |
| Assembled Scaffolds
help
|
1 | Assembled Scaffold Length (bp)
help
|
103552 |
Tezzeta Chloroplast Genome
| Total Sequenced Bases (bp)
help
|
4,398,963 | Mean Quality
help
|
10.0 |
| Mean Read Length (bp)
help
|
910.8 | N50 Read Length (bp)
help
|
1,295.0 |
| Proportion of Chloroplast Bases (%)
help
|
5.1 | Chloroplast Coverage
help
|
1 |
| Assembled Contigs
help
|
0 | Assembled Contig Length (bp)
help
|
0 |
| Assembled Scaffolds
help
|
0 | Assembled Scaffold Length (bp)
help
|
0 |
Daffodil Phylogeny
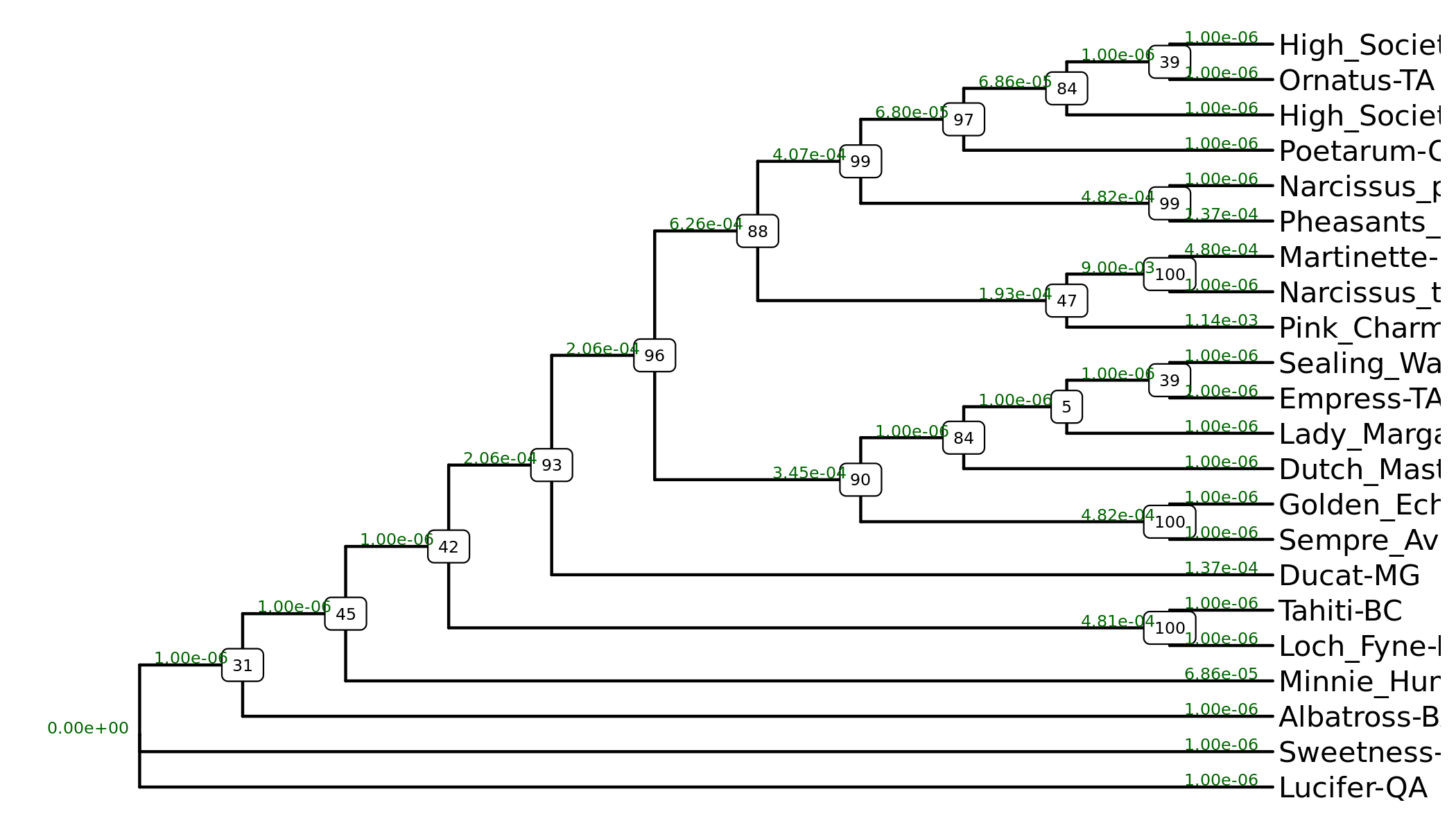
Genomic Multiple Sequence Alignments
Two multiple sequence alignments have been carried out. The first includes all (including partial) sequences, while the second includes just sequences > 120kb in length, which were used for carrying out the phylogenetic analysis.
The JalviewJS link below for the 'long' sequences will also load they phylogenetic tree in addition to the alignment.
Phylogeny help

Protein Multiple Sequence Alignments
Each identified gene within the assembled chloroplast genomes has been translated to a protein sequence, and then a multiple sequence alignment generated for each gene. Alignments were created using Muscle 5.1 with default parameters.