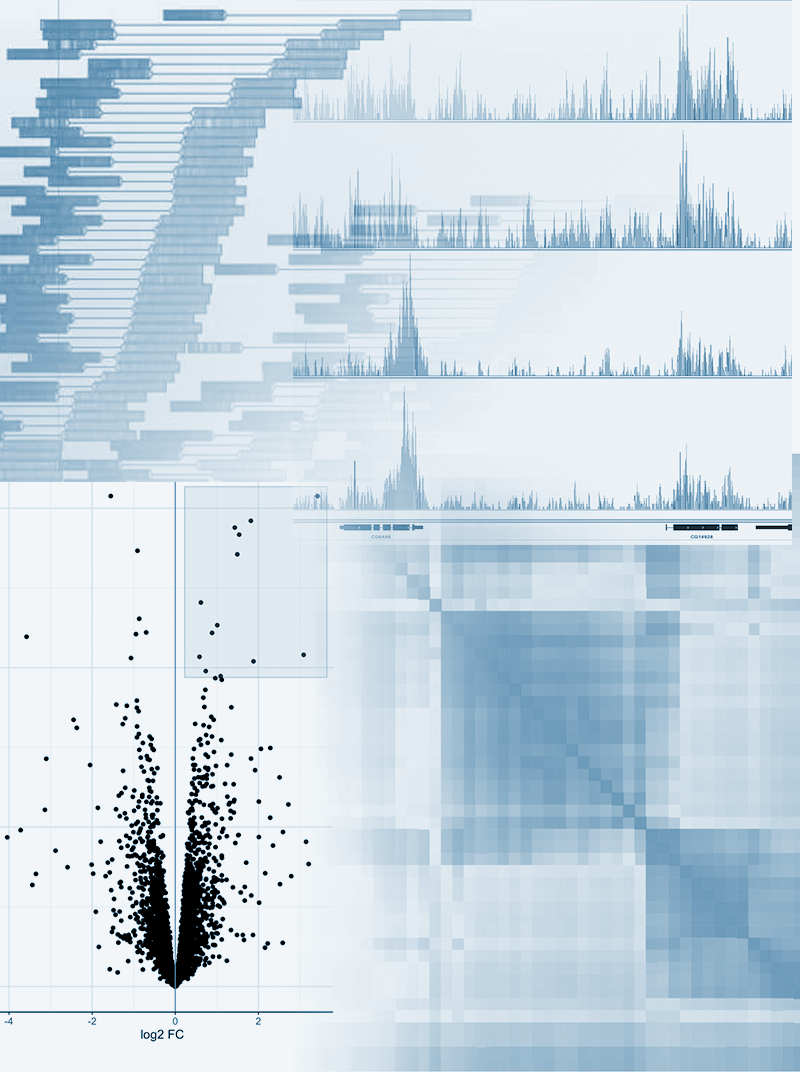
Module Overview
ChIP-Seq allows the investigation of proteins interacting with DNA though the immunoprecipitation of bound proteins, producing a library of DNA molecules bound to the protein of interest. Sequencing of these fragments reveals the location of the binding sites within the genome. This module will discuss ChIP-Seq experimental design, identification of peaks in read alignments indicating binding loci and differential peak calling between experimental conditions.
Learning Outcomes
After completing this module, participants will be able to:
- Design reproducible ChIP-Seq experiments
- Understand the different type of signals resulting from different kinds of binding interaction
- Identify peaks in read alignments resulting from protein binding
- Identify differential binding between experimental conditions
Prerequisite Modules/Knowledge
- Introduction to Linux
- The HPC Cluster
- Introduction to NGS
- Read Alignment
Course Schedule
Not currently scheduled
Join Waiting List